Genetic Variation Exploration in Non-Model Organisms with OmicsBox
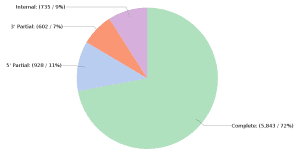
The study of genetic variation is fundamental in genomics research as it enables the comprehension of mutation causes and effects in the diversity of life. However, research in this field is well-developed in model organisms, while non-model organisms lack sufficient guidance for a consistent variant analysis pipeline. OmicsBox is a robust tool for investigating genetic variation in non-model organisms, facilitating