Input Reads
1
M
high Quality transcripts (BUSCO)
1
%
ASSEMBLY TIME
1
h
The OmicsBox Transcriptomics module allows you to process RNA-seq data from raw reads down to their functional analysis in a flexible and intuitive way.
Use FastQC and Trimmomatic to perform the quality control of your samples, to filter reads and to remove low quality bases.
Assemble short reads with Trinity to obtain a de-novo transcriptome without a reference genome. Assess the completeness of the transcriptome with BUSCO and cluster similar sequences with CD-HIT. Moreover, you are able to predict coding regions with TransDecoder or assess the coding potential of each sequence with CPAT.
Align RNA-seq data to your reference genome making use of STAR (Spliced Transcripts Alignment to a Reference) or BWA (Burrows-Wheeler Aligner) regardless of your hardware. In addition, BAM-QC provides several useful modules to evaluate RNA-seq alignment files.
Quantify expression at gene or transcript level through HTSeq or RSEM and with or without a reference genome.
Detect differentially expressed genes between experimental conditions or over time with well-known and versatile statistical packages like NOISeq, edgeR or maSigPro. Rich visualizations help to interpret results.
Use LongQC to assess the quality of long-read datasets without a reference genome.
Identify long-read-sequenced transcripts with IsoSeq3, FLAIR, or IsoQuant and then perform a long-read transcriptome analysis and characterization using SQANTI3. With this implementation, you will obtain a curated transcriptome including a detailed analysis report.
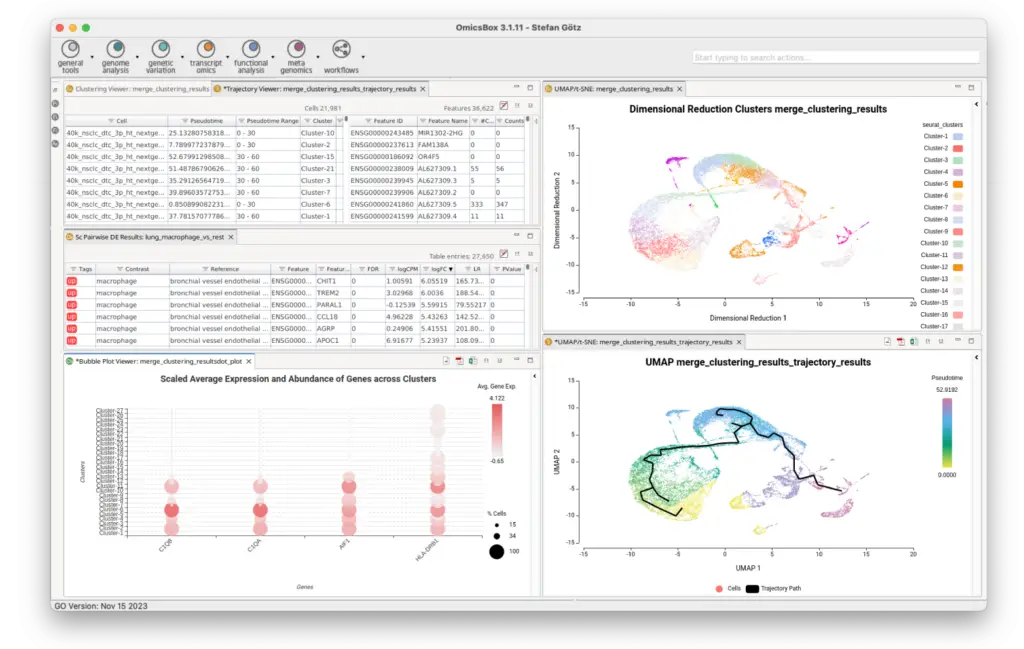
Obtain scRNA-Seq counts seamlessly with STARsolo for different library-prep technologies. Perform Single-Cell RNA-Seq clustering with Seurat to identify groups of cells and examine marker genes’ expression. Gain insight into cell transitions with Monocle3 and visualize cell lineage trajectories in pseudo-time.
By combining differential expression results with functional annotations, enrichment analysis allows to identify over- and underrepresented biological functions.
Unlock the power of advanced Single-Cell insights with OmicsBox.
With our user-friendly platform, Single Cell RNA-Seq analysis becomes accessible to every scientist, with a focus on dynamic data exploration and interactive visualizations.
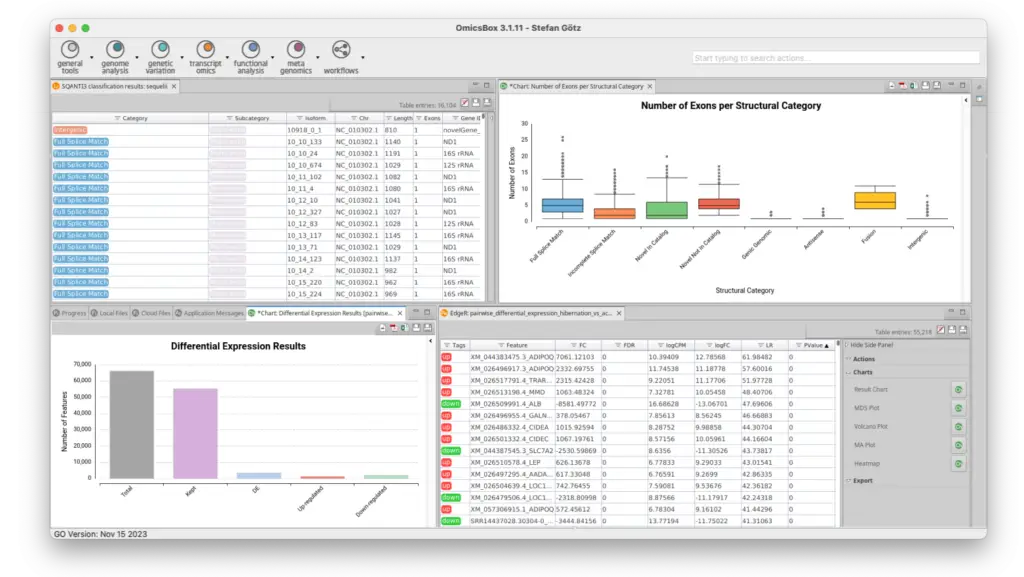
Characterize and Curate Long-Read Transcriptomes with OmicsBox.
Discover our set of long reads tools to accurately reconstruct a transcriptome, assess the quality, and quantify isoforms easily.
Get familiar with all new Modules and Features with a Free Trial or a Custom Demo.
OmicsBox works out of the box on any standard PC or laptop with Windows, Linux and Mac.
Generate your own reference transcriptome by de novo assembling RNA-seq reads. Assess the completeness of the assembly, cluster similar sequences to reduce redundancy. Finally, predict coding regions and find homologous sequences to characterize transcript sequences.
