Support
Professional Bioinformatics Support
Our support and development team works closely together to take care of all the issues you might have, doesn’t matter if rather technical, about bioinformatics or biology.
You can reach us a support@biobam.com and we will do our best to answer all requests within one day. All comments and suggestions regarding our services are most welcome and a valuable source of information for us.
To share your experiences or to search for a specific solution within OmicsBox please log in to your BioBam Account here.
Thank you in advance for any future contributions.
BioBam Help Center
Our Help Center will answer all your questions about OmicsBox. Learn how to use it with our extensive user manual, datasets and much more.

White Papers
Download white papers, product sheets and brochures.
- OmicsBox
- OmicsCloud
- Blast2GO Command Line Tools
- Product Sheet
Teaching Material
Use OmicsBox to explain basic bioinformatics concept like sequence alignments with BLAST, functional annotation with the Gene Ontology, public biological databases, NGS data analysis pipelines, etc.
If you are planning a teaching activity, please let us know and we may provide you with teaching presentations, and practical sessions including exercises with solutions and datasets.
Please write to us: support@biobam.com
We are here to help!
If you have any comments, suggestions or whatever trouble with OmicsBox, Blast2GO or any other product or service and want to get directly in touch with us, please write an e-mail to support@biobam.com
OmicsBox Webinars

33:18

35:53
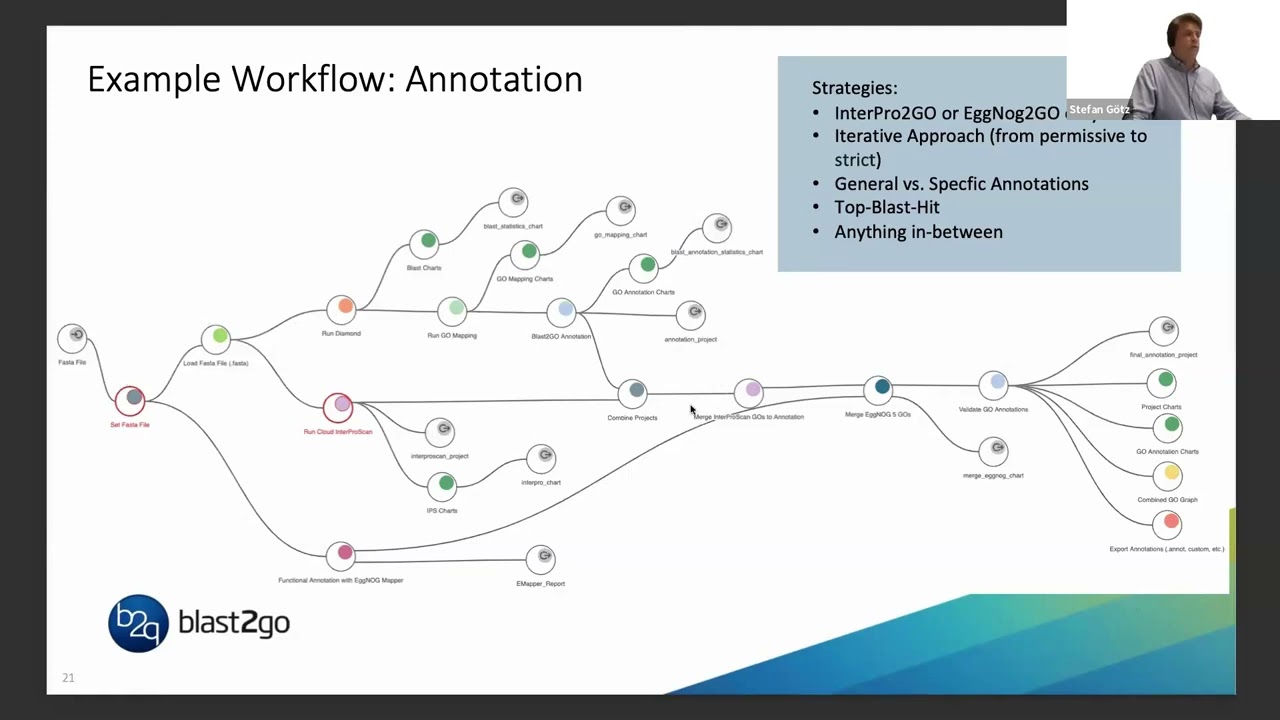
56:40
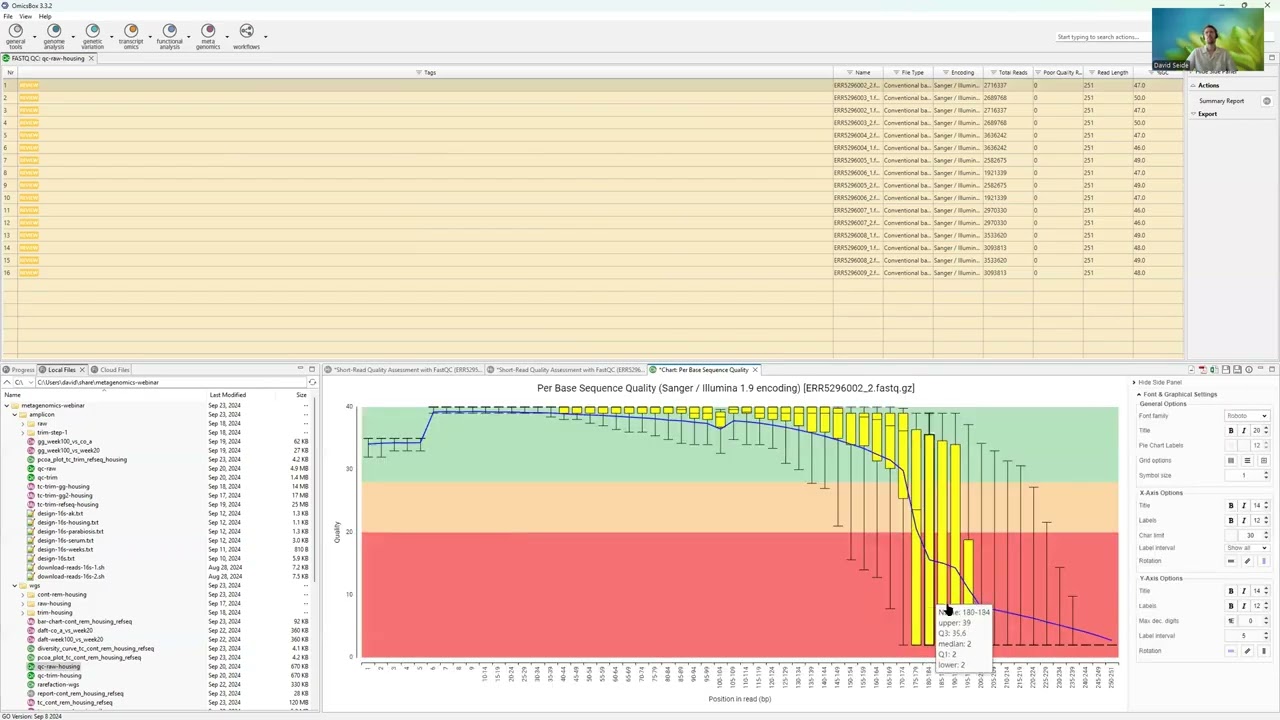
46:19
Video Library
-

OmicsBox: The All-In-One Bioinformatics Software
-

Single-Cell RNA-Seq Clustering with OmicsBox 2.2
-

Long Read Quality Assessment with LongQC
-

Transcriptome Analysis with SQANTI3 in OmicsBox 2.2
-

scRNA-Seq Trajectory Interference with Monocle 3
-

Single-Cell RNA-Seq Differential Expression Analysis with OmicsBox 2.2
-

How to use the MLST App in Omicsbox
-

OmicsBox Presentation
-

How to use "Retrieve Blast Top-Hit" with OmicsBox/Blast2GO



