Fast Variant Calling
Choose between two of the best algorithms to find variants in your alignment files: BCFtools mpileup and Freebayes. Using OmicsBox, not only you will receive a VCF file, but also you will be able to visualize different charts to control the quality of your results.
Model & Non-Model Variant Annotation
Gather information about the coding and genetic consequences of all the variants included in a VCF file with the Variant Effect Predictor of Ensembl. You will only need the reference genome used in the Variant Calling step and the annotation file.
End-to-end Analysis
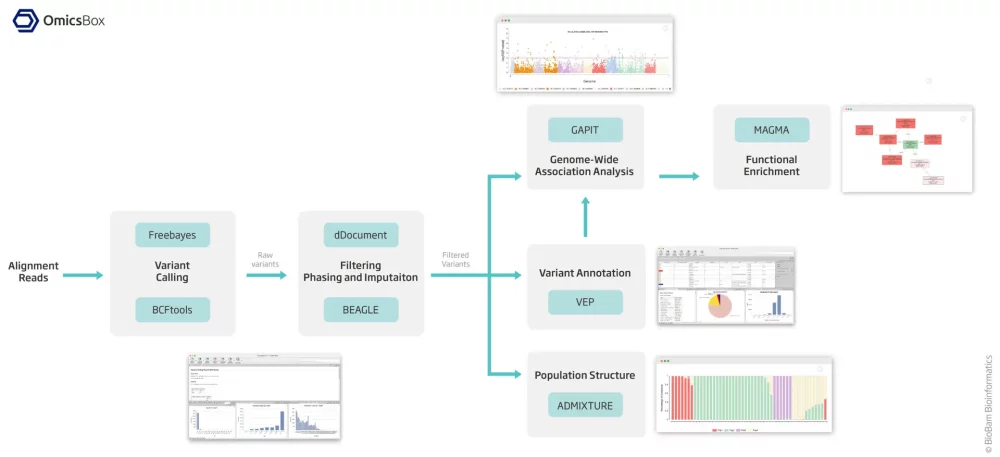
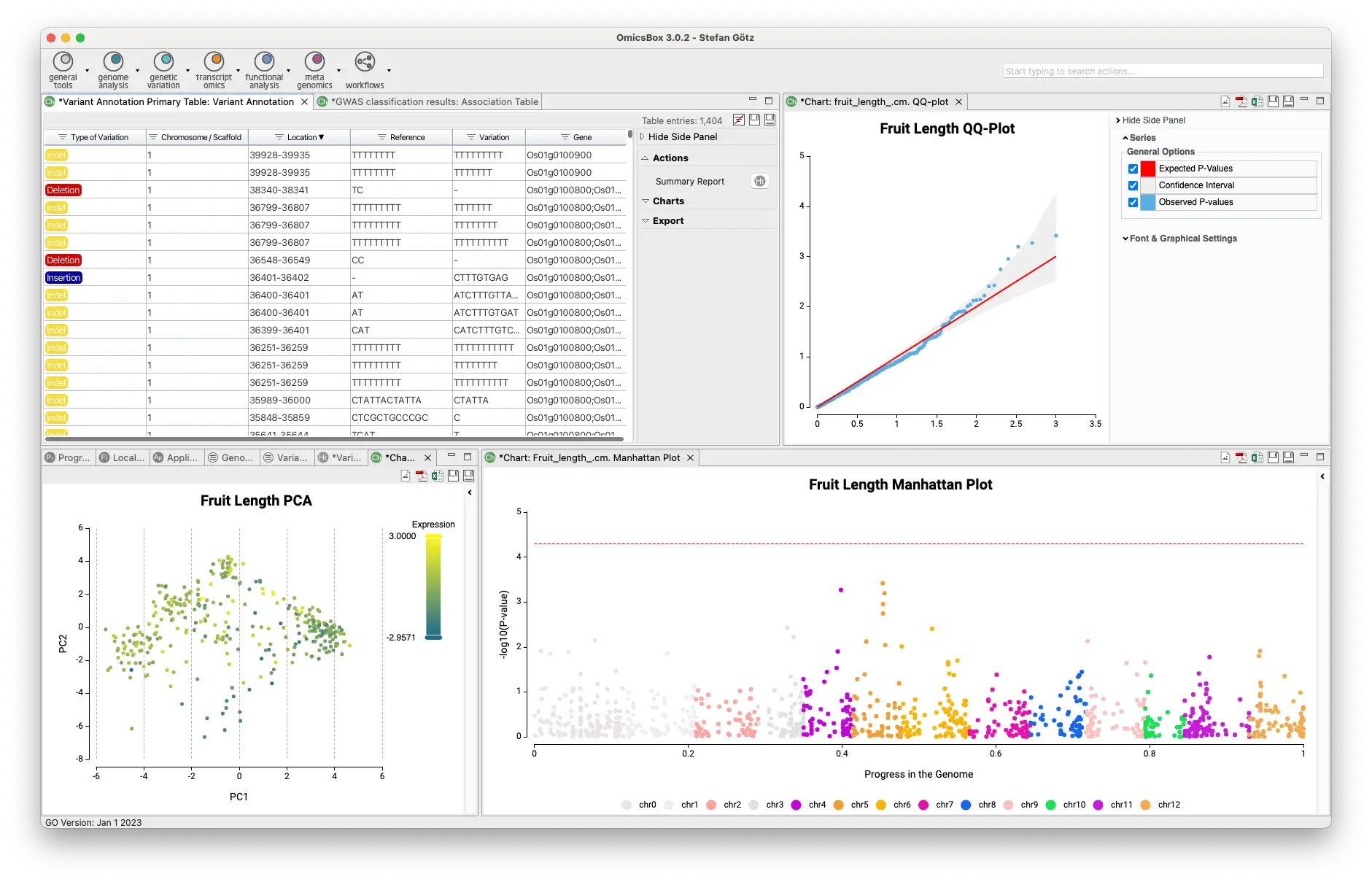
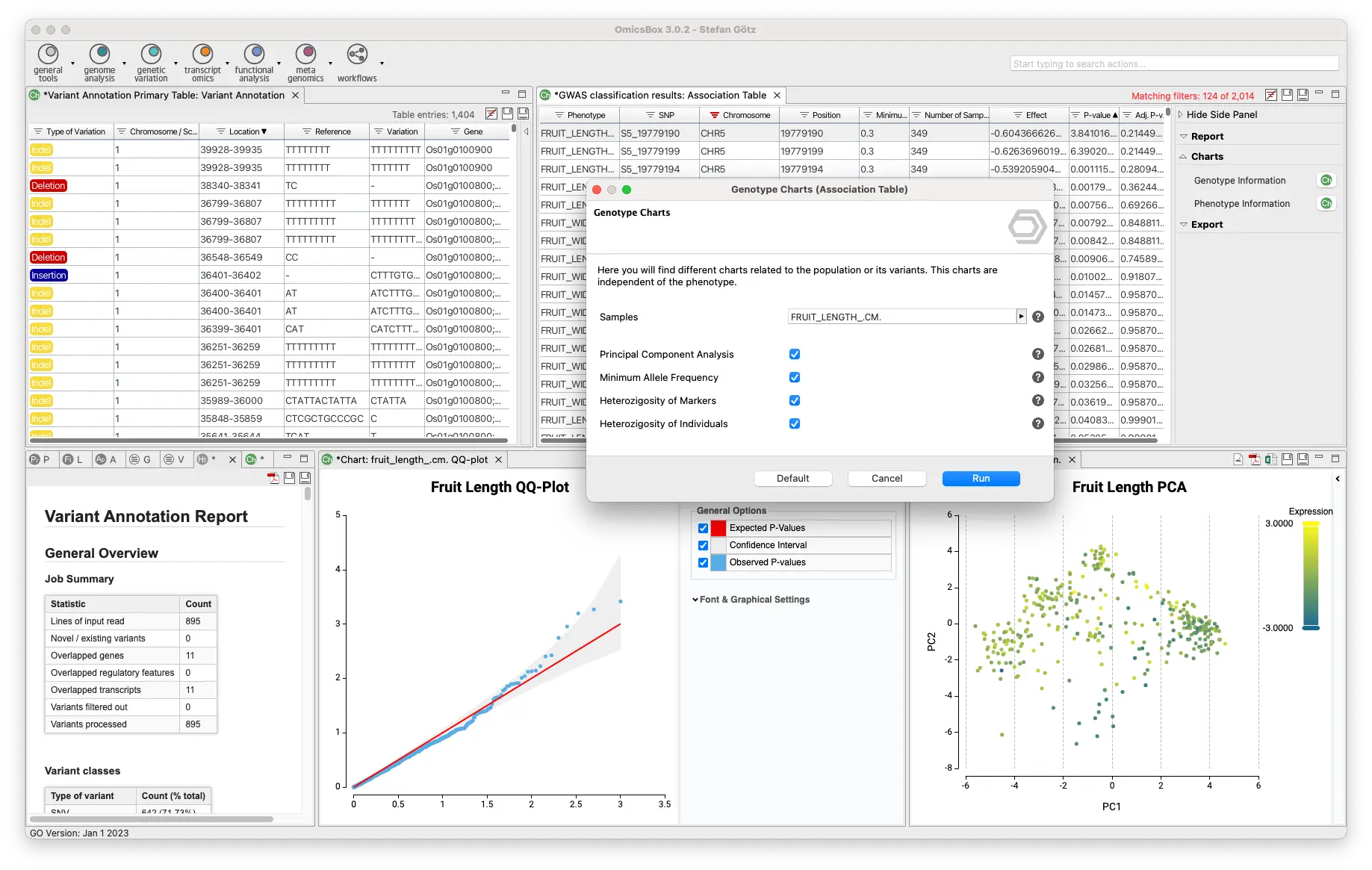
Create your VCF files with a Variant Calling algorithm, merge or extract information, and filter your VCF files. Then, you will be able to phase and impute your variants and perform a downstream analysis such as GWAS or Population Structure Analysis.