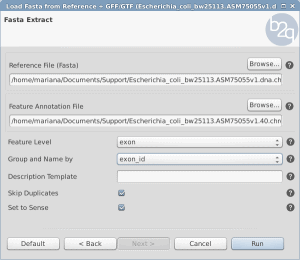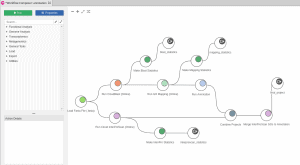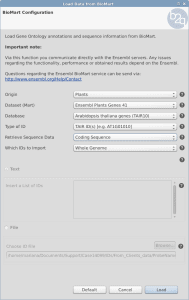
Load Sequences and Annotation from a list of identifiers
OmicsBox/Blast2GO offers two different features to retrieve the gene/protein sequences as well as the corresponding annotation from a list of identifiers within Blast2GO PRO. Both features can be found under File > Load > Load Annotations. The expected input file is a text file with the identifiers in a single column without a header.