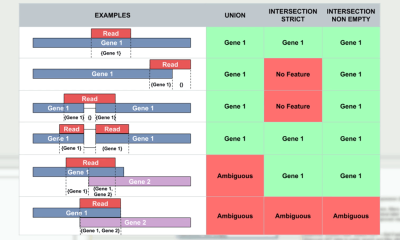
The “Create Count Table” feature of OmicsBox/Blast2GO allows quantifying the gene expression of RNA-seq datasets.
This video shows step-by-step how to create a count table of raw reads and explains in detail different concepts of expression quantification. The available parameters are inspired by the popular HTSeq Python Package. (reference below).
As input, aligned sequencing reads in SAM/BAM format and a GTF/GFF file with coordinates of genomic features are required. The output, an un-normalized count table, can then be analysed directly within Blast2GO. Various options for differential expression analysis are available (find videos here and here).
Reference:
Anders S, Theodor Pyl P, Huber W (2015). “HTSeq — A Python framework to work with high-throughput sequencing data.” Bioinformatics, 31 (2), p. 166-169.