Metagenomic analysis of two soda lakes, with and without cyanobacterial bloom, with OmicsBox
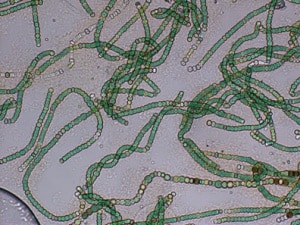
In this use case we will use the metagenomics tools included in OmicsBox to analyze the microbial communities of two different soda lakes from Brazil. The original study was carried out by Ana P. D. Andreote, et al., 2018 (doi: 10.3389/fmicb.2018.00244). Introduction Soda lakes are special ecosystems found across Africa, Europe, Asia, etc. These lakes show high levels of sodium