In the field of bioinformatics and genomic research, data representation is a critical aspect that influences the ease and accuracy of analysis. Two common file formats used in genomic data representation are Gene Transfer Format (GTF) and General Feature Format (GFF). While they may seem similar at a glance, there are distinct differences between these two formats that are crucial for researchers to understand. This post delves into the scientific nuances of GTF and GFF files, examining their structure, usage, origins, and differences.
GFF and GTF files are widely used in various aspects of genomic research due to their ability to represent complex genomic data in a standardized format. GFF files, with their flexibility and broader scope, are commonly used in genome mapping, annotation, and comparative genomics. They provide a comprehensive overview of genomic features across DNA, RNA, and proteins.
GTF files, being more gene-centric and detailed in gene structure representation, are particularly useful in gene expression studies such as RNA sequencing (RNA-Seq) and transcriptome analysis. Their structured format facilitates accurate mapping and quantification of gene expression levels, making them a vital tool in functional genomics and gene regulation studies.
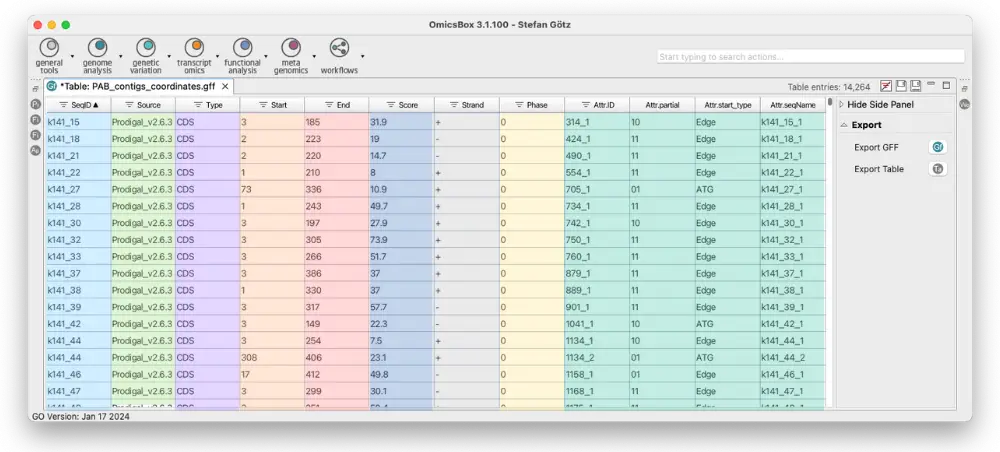
| Column | Name | Description |
| 1 | SeqID | The name of the sequence where the feature is located. |
| 2 | Source | The algorithm or procedure that generated the feature. This is typically the name of a software or database. |
| 3 | Type | The feature type name, like “gene” or “exon”. In a well-structured GFF file, all the children’s features always follow their parents in a single block (so all exons of a transcript are put after their parent “transcript” feature line and before any other parent transcript line). In GFF3, all features and their relationships should be compatible with the standards released by the Sequence Ontology Project. |
| 4 | Start | Genomic start of the feature, with a 1-base offset. This is in contrast with other 0-offset half-open sequence formats, like BED. |
| 5 | End | Genomic end of the feature, with a 1-base offset. This is the same end coordinate as it is in 0-offset half-open sequence formats, like BED. |
| 6 | Score | Numeric value that generally indicates the confidence of the source in the annotated feature. A value of “.” (a dot) is used to define a null value. |
| 7 | Strand | A single character indicates the strand of the feature. This can be “+” (positive, or 5′->3′), “-“, (negative, or 3′->5′), “.” (undetermined), or “?” for features with relevant but unknown strands. |
| 8 | Phase | Phase of CDS features; it can be either one of 0, 1, 2 (for CDS features) or “.” (for everything else). See the section below for a detailed explanation. |
| 9 | Attributes | A list of tag-value pairs separated by a semicolon with additional information about the feature. |
Background: The Importance of File Formats in Genomics
File formats in genomics, like GTF and GFF, standardize the representation of genomic information, ensuring consistency and compatibility across different computational tools and databases. Efficient representation of complex genomic data is crucial for computational tools to accurately interpret and analyze genetic information.
The General Feature Format (GFF)
GFF is a standard file format used for storing genomic sequences and annotations, developed by the Sanger Centre (v2) and the Sequence Ontology Project (v3). It describes features and annotations of DNA, RNA, and protein sequences in a plain text format with nine fields separated by tabs.
The Gene Transfer Format (GTF)
GTF, often seen as a derivative of GFF, is more specific in its application. It was generally termed GTF around 2000 and developed as part of collaborative genome annotation projects. GTF borrows from GFF but has additional structures for detailed gene annotations.
Please Note: Since GFF version 2, the columns and format of both file types are the same.
- Specificity in Annotation: GTF is more gene-centric, while GFF is broader in genomic feature representation.
-
Attribute Column Format: The attribute column in both formats is typically structured as a series of key-value pairs (separated by semicolons) and contains meta-information such as gene or transcript names or IDs, biotype, etc. The attribute column in GTF often includes specific tags like gene_id and transcript_id. The GFF’s attribute column is often more flexible.
-
Intended Use and Focus: GTF is used to describe the structure of genes, whereas GFF is used for a broader range of genomic annotation tasks (i.e. annotating genomic features).
-
Compatibility with Bioinformatics Tools: Certain bioinformatics tools are specifically tailored to work with either GTF or GFF due to their structural differences.
Conversion Between GTF and GFF
Conversion between these formats is possible with various tools and scripts, but caution must be exercised to ensure data integrity.
Two popular tools for the conversion between GFF and GTF file formats are:
-
AGAT: This is a bioinformatics tool that provides a comprehensive suite of scripts to manipulate and analyze genome annotation files. It includes functionality for converting between GFF and GTF formats.
-
GenomeTools: This is a versatile software package for bioinformatics, offering a wide range of tools for genomic data analysis. Among its various capabilities, it includes tools for converting files from GFF to GTF format and vice versa.
Both of these tools are widely used in the bioinformatics community for their robustness and utility in handling genomic file formats.
Historical Context and Development
-
GFF Development: The GFF format, with its initial versions developed by the Sanger Centre and later versions by the Sequence Ontology Project, addressed the need for a general and flexible format for genomic feature representation. GFF3, in particular, was developed to overcome the limitations of GFF2, such as its inability to represent a three-level hierarchy of gene, transcript, and exon.
-
GTF Development: The GTF format emerged around 2000 from the GFF format to meet the specific needs of gene-centric annotations in various genome annotation projects.
Conclusion
Understanding the differences between GTF and GFF files is crucial in genomic data analysis. Researchers must carefully choose the appropriate file format based on their specific research needs to ensure the accuracy and efficiency of their analyses.
Useful Links
-
Gene Transfer Format (GTF) on AGAT Documentation
References
- “General feature format.” Wikipedia, The Free Encyclopedia.
- The GTF/GFF formats. AGAT Documentation.
About the Author
 Stefan Götz
Stefan started his career in computer science and developed a keen interest in biomedical applications. He transitioned from biomedical informatics to computational biology, specializing in functional genomics and sequence analysis. After completing a Ph.D. in bioinformatics, Stefan chose to pursue a non-academic path and became an entrepreneur. In 2011, he founded BioBam, a bioinformatics company aimed at advancing genomics research to enhance human health, food safety, and environmental quality.
As the CEO of BioBam, Stefan is responsible for various aspects of the company's growth, such as business strategy, product management, team leadership, and research and innovation.
Stefan Götz
Stefan started his career in computer science and developed a keen interest in biomedical applications. He transitioned from biomedical informatics to computational biology, specializing in functional genomics and sequence analysis. After completing a Ph.D. in bioinformatics, Stefan chose to pursue a non-academic path and became an entrepreneur. In 2011, he founded BioBam, a bioinformatics company aimed at advancing genomics research to enhance human health, food safety, and environmental quality.
As the CEO of BioBam, Stefan is responsible for various aspects of the company's growth, such as business strategy, product management, team leadership, and research and innovation.