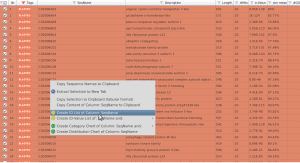
How to create a gene list within OmicsBox to run the functional enrichment analysis
This article explains how to create ID lists of either Blast results or differential expressed genes in Blast2GO.
Mini-tutorials for common use-cases and to address frequently asked questions FAQs

This article explains how to create ID lists of either Blast results or differential expressed genes in Blast2GO.
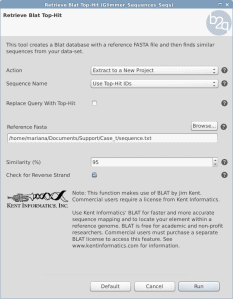
When running GeneFinding the sequences receive a name with the predicted genes. The first part of the sequence identifier comes from the genome reference sequence name (de-novo assembly) and then a _orfx is appended, where x is a number. Sometimes this name is not useful to proceed with downstream analysis or compare results from other experiments. Is there any way
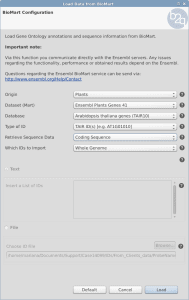
OmicsBox/Blast2GO offers two different features to retrieve the gene/protein sequences as well as the corresponding annotation from a list of identifiers within Blast2GO PRO. Both features can be found under File > Load > Load Annotations. The expected input file is a text file with the identifiers in a single column without a header.
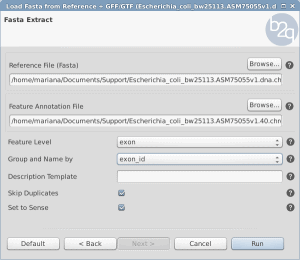
Sometimes databases provide the whole genome and the GFF or GTF files but not the exon or CDS FASTA files.With OmicsBox/Blast2GO it is possible to load a Fasta sequences and to extract the exons or the CDS from the genome using the GFF file.
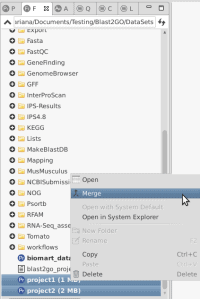
OmicsBox allows combining two Functional Anlaysis projects which contain either have the same identifiers or different ones.
CloudBlast and CloudIPS need Cloud Units to work.If all Cloud Units have been consumed CloudBlast and CloudIPS will stop. Consumption depends on what you are doing. The most costly (in terms of computation time) analysis is definitely to do a blastx against the whole NR with very long sequences. The smaller the database used the less Cloud Units are used. Use the taxonomy
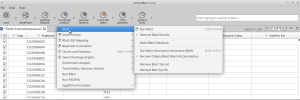
After loading the sequences, the run blast icon is greyed out in OmicsBox, why? It is possible to load more than one project into OmicsBox and each project will be displayed in a different tab.So to run the analysis on the different projects you will need to have the OmicsBox table active (white tab). In your case, probably the Progress
It is possible to perform a Fisher Exact Test on the 2 groups even if the annotation is in different files (.annot). First, both .annot files (group 1 and group 2) need to be loaded into Blast2GO.The test and reference set have to be generated according to the groups you want to compare. These are normal text files with the

This article explains what a Gene Set Enrichment Analysis (GSEA) is, how it works and how to use it with OmicsBox. What is an enrichment analysis? Enrichment analyses are a family of bioinformatics methods that aim to facilitate the biological interpretation of many bioinformatics results. Among these, the methods based on gene sets analysis are particularly helpful and widespread. The

