OmicsBox/Blast2GO offers the option to create either single or ranked ID lists depending on the algorithm that will be used for the functional enrichment analysis, the Fisher Exact Test or GSEA.
On one hand, the Fisher Exact Test expects a single column with the sequence identifiers, which have to match the ones with the Blast2GO annotation project.
On the other hand, the GSEA needs a ranked list, where the first column is the sequence identifier and the second one could be e.g the logFC, P-Value of a differential expression project.
Use cases
1 – ID list of blasted sequences
In a study, one may want to know which genes are pathogenic.
Here, one can run a blast search against a Putative Pathogen-Host Interaction (PHI) Genes database and the sequences retrieved from blast results can be used as the list of interest.
With the Blast2GO project opened, there is the need to, first, filter by the blast (orange sequences) and then create the ID list.
Filter out sequences and create ID list:
It is very easy to filter out those sequences that do not have blast results within Blast2GO.
- Click the document on the Tags column header.
- Check the Blasted checkbox (Figure 1).
- Only the Blasted sequences will be shown in the table (Figure 2).
- Mark all sequences with Ctrl + A (Windows and Linux) / Apple + A (Mac).
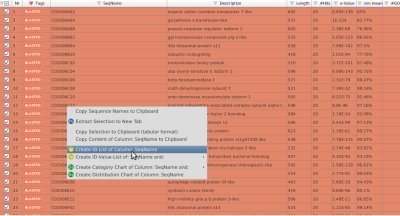
- Right-click on the table under SeqName and in the context menu choose to Create ID List of Column: SeqName (Figure 3).
- A new tab will open and the list of interests will be generated.
- Save the lists as a b2g project (go to File > Save).
- Use the .b2g in Fisher’s Exact Test as a test set (Analysis > Enrichment Analysis).
2 – ID list from Time Course Differential Expression analysis
Since the time course expression analysis classifies genes according to their expression over time, it is possible to use this classification to extract subsets of interesting genes.
This subset can be used as a test set for Fisher’s Exact Test. Gene subsets can be extracted from a Time Course Expression Project according to two criteria: expression behavior over time or clusters.
Filter out genes and create ID list:
- The time-course expression analysis labels each significant gene according to the expression pattern it shows over time. For instance, The START tag means that the gene shows a significant expression at the start of the experiment (the meaning of each tag is explained in the summary report and also in the user manual). You can extract a subset of genes that are labeled with a specific tag:
- Filter the table by the Tags columns. It is possible to specify one or more tags.
- Mark all filtered entries by Ctrl+A (Windows and Linux) or Apple + A (Mac).
- Right-click on the SeqName column and click on the Create ID list of columns: SeqName option in the context menu. A new tab will open with the ID list of the filtered entries.
- Save the lists as a b2g project (go to File > Save).
- This .b2g object can be used as a test set for the Fisher’s Exact Test.
- On the other hand, the time-course expression analysis returns clusters of genes. Genes belonging to the same group have a similar expression pattern over time. To obtain the list of genes belonging to a specific cluster, open the Summary Report and go to the Profile Clusters section (Figure 4).
- Click on the Id button to create an ID list of the genes of the cluster. This list can be saved and used as a test set for the Fisher’s Exact Test.
Note: In both scenarios, to run the Fisher Exact Test an annotated Blast2GO Project is needed and the identifiers from the project and the ID lists must match to each other.