Decoding Long-Read Sequenced Transcriptomes: FLAIR vs. StringTie2
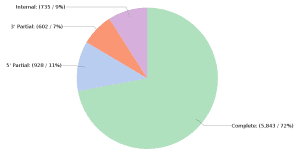
Transcriptome reconstruction is a challenging bioinformatic problem. The development of long-read sequencing technologies has made it easier to solve this issue thanks to the possibility of having transcripts entirely contained in a read. In addition, different algorithms have emerged to generate transcriptomes from long-read datasets, such as FLAIR and StringTie2. Both tools can be used for transcriptome reconstruction with long-read data.