
Whole Genome Functional Annotation of Solanum lycopersicum
Whole genome functional annotation of Solanum lycopersicum with OmicsBox functional annotation module.

Whole genome functional annotation of Solanum lycopersicum with OmicsBox functional annotation module.

We attended PAG XXVII, the largest Ag-Genomics Meeting in the world. https://www.intlpag.org/30/ We launched OmicsBox Beta and gave away a whole bunch of free subscriptions. Here are a few pics of the happy winners! Hopefully, they took advantage. The PAG 2019 Team with our new OmcisBox Booth :-). See you next year!
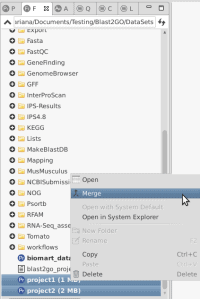
OmicsBox allows combining two Functional Anlaysis projects which contain either have the same identifiers or different ones.

Blast2GO Supported Project. Researchers: MSc Sara D. Cardoso, Ph.D. candidate at Gulbenkian Institute for Science (IGC), Oeiras, Portugal Supervisors: Prof. Rui F. Oliveira, Gulbenkian Institute for Science (IGC), Oeiras, Portugal and Prof. Adelino V. M. Canário, CCMAR – Centre of Marine Sciences, University of Algarve, Faro, Portugal Background and Project Overview: The peacock blenny Salaria pavo (family Blenniidae) is a
Release Date: 13/01/2018) Just in time for the PAG Plant & Animal Genome Conference, we launched Blast2GO 5! This major release brings many new features. Most importantly, Blast2GO is now connected to the powerful BioBam Cloud, an AWS architecture which gives a big boost to many analysis steps. Details below. To upgrade from version 4 to 5 you have to

BioBam Supported Project: Changes in maize transcriptome in response to Maize Iranian Mosaic Virus (MIMV) infection. Researchers: Mr. Abozar Ghorbani, Ph.D. candidate at Plant Virology Research Center, College of Agriculture, Shiraz University, Shiraz, Iran Supervisors: Prof. Keramatollah Izadpanah, Plant Virology Research Center, College of Agriculture, Shiraz University, Shiraz, Iran and A/Prof. Ralf Dietzgen, Queensland Alliance for Agriculture and Food Innovation, the

The OmicsBox Genome Analysis Module allows executing eukaryotic de-novo and RNA-seq based gene finding with Augustus. In this way, it is possible to discover novel, putative coding genes and their genomic positions for yet uncharacterized genome. Based on the Augustus algorithm an ‘ab-initio’ (DNA sequences only), as well as RNA-seq guided (BAM files) gene predictions, are supported. We will
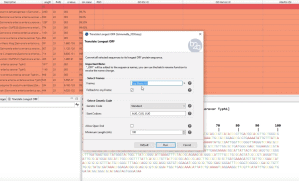
The Blast2GO “Translate Longest ORF” tool searches for the longest Open Reading Frame (ORF) in nucleotide sequences and translates them into their protein sequences. You may choose one or multiple of the six possible DNA frames or select the reading frame based on the frame of the best blastx hit. In this video, we will explain how to translate a set of

(Release Date: 13/09/2016) We are very happy to announce the release of Blast2GO 4. This new version contains many new bioinformatic features like Differential Expression Analysis, Gene Finding or Gene Set Enrichment Analysis. Blast2GO will update automatically if activated. Otherwise please download the latest version online from here. Feedback, questions, as well as feature requests, are most welcome. Please write
