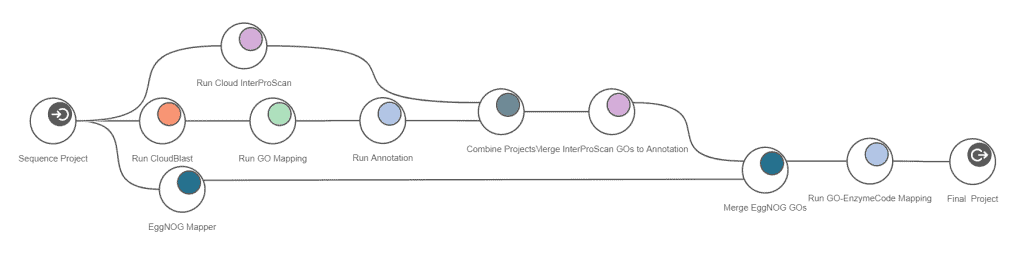
OmicsBox with the Blast2GO functional annotation module is a high-throughput solution for functional annotation of novel sequence data to obtain high-quality functional labels. Functional annotation is relevant to complete the functional characterization of de novo sequenced genomes and transcriptomes. This application note describes the complete functional annotation of the tomato genome, Solanum lycopersicum, using the functional annotation module in OmicsBox. We present the basic analysis workflow, describe several issues regarding the quality and quantity of the obtained results, and comment on the genome wide analysis of the functional compositions of this dataset. The complete analysis was carried out in just 4 hours.


Data
In the following analysis, we use a whole genome dataset of Solanum lycopersicum (Taxa: 4081). The official CDS annotation for the genome is obtained from the SL2.40 genome build released by the International Tomato Annotation Group (Release 3.2, 2017-06-15). ftp://ftp.solgenomics.net/tomato_genome/annotation/ITAG3.2_release/ITAG3.2_CDS.fasta
Sequence Alignment via BLAST
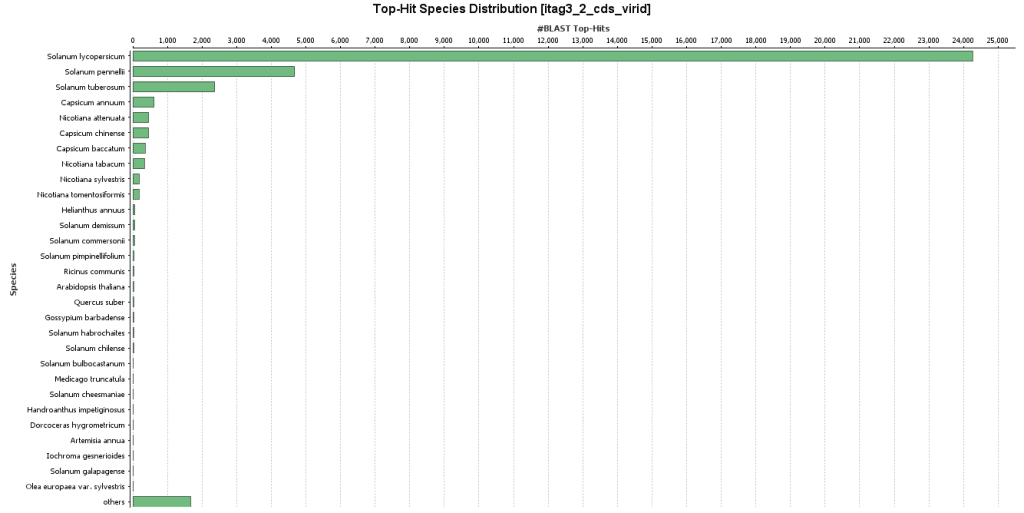
First, the tomato CDS sequences are used as queries in a blastx search launched with CloudBlast. The BLAST search is run against the Viridplantae subset of the non-redundant (NR) database with blastx-fast, an e-value of 1xE-6 and we keep the top 20 alignments for each sequence.
Domain Search via InterProScan
An InterPro domain search is performed via CloudInterProScan. InterPro combines different protein signature recognition methods and the identified domains can be directly translated into Gene Ontology terms. InterProScan and BLAST searches can be performed in parallel and then combined.
Orthologous Group Search with EggNOG-Mapper
The eggNOG database is a database of biological information hosted by EMBL. It is based on the original idea of COGs (clusters of orthologous groups) and expands that idea to non-supervised orthologous groups constructed from 2000 organisms in version 4.5.1. EggNOG-mapper is a tool for fast functional annotation of novel sequences (genes or proteins) using precomputed eggNOG-based orthology assignments. The use of orthology predictions for functional annotation avoids transferring annotations from paralogs e.g. duplicate genes with a higher chance of being involved in functional divergence.
Gene Ontology Mapping
GO mapping is the process by which the Blast2GO methodology retrieves functional information for all BLAST Hits from the Gene Ontology (GO) database. The GO database contains several millions of functionally annotated gene products for hundreds of different species. OmicsBox uses different public resources provided by the NCBI, PIR and GOA to link the different protein IDs (names, symbols, accessions, UniProts, etc.) to these GO annotations. Moreover, annotations in the GO database contain an evidence code qualifier that provides information about the quality of this functional assignment, which is also retrieved by OmicsBox. The result of this step is a set of GO candidate annotation terms for each tomato query sequence.
Functional Annotation
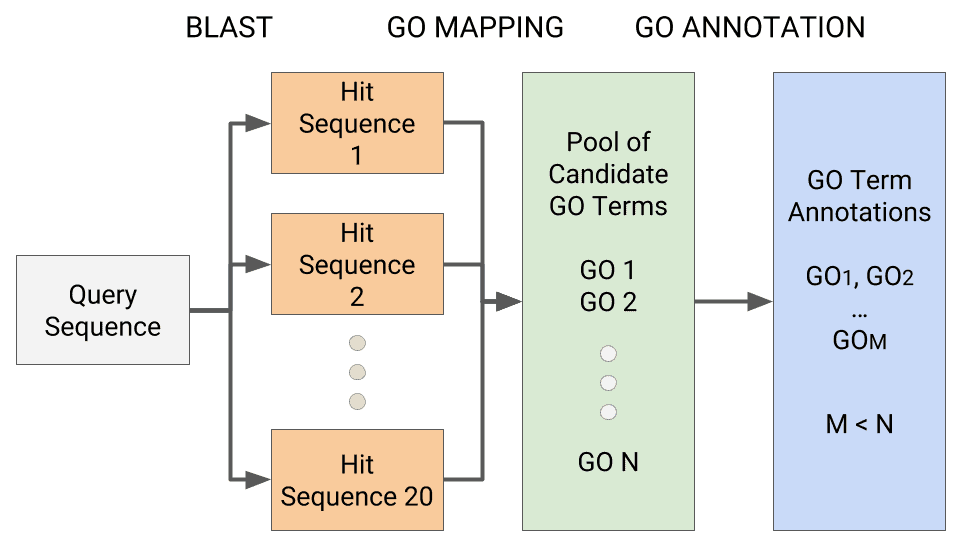
The annotation algorithm selects GO terms from the pool of candidate GOs obtained by the GO mapping step and assigns these to the query sequences. GO annotation is carried out by applying the Blast2GO annotation rule, which computes an annotation score for each candidate GO term. This score considers the similarity between hit and query sequences, the evidence code of each GO term and the existence of neighbouring GO term candidates. We use an annotation threshold of 55 to select the GO term candidates for final assignment to the query sequence, leaving other parameters by default to achieve an optimal balance between quality (minimum of 55% of sequence similarity for hits with experimental evidence codes, higher for other types of evidence) and quantity (number of annotated sequences). The Blast2GO annotation rule is described in S. Götz et al., 2008. Figure 1 represents schematically the Blast2GO annotation process.
Combined Graph and Pie Chart
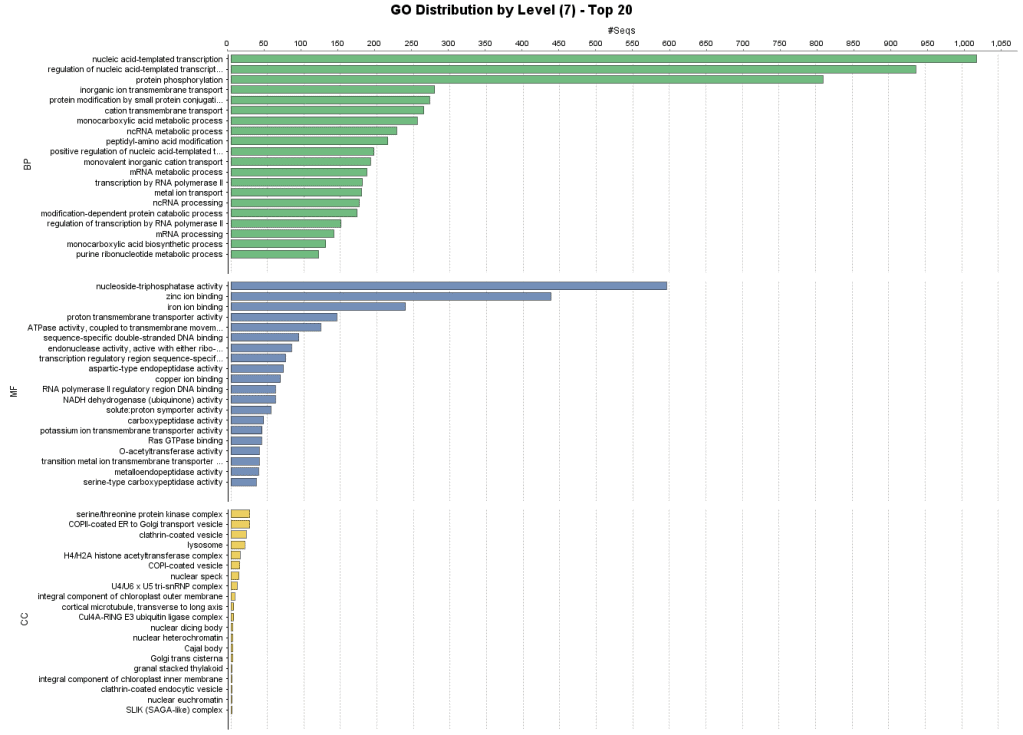
Once we reduce the functional diversity to an appropriate size, we can obtain a Combined Graph. The Combined Graph provides a bird’s eye view of the GO terms annotations of the entire dataset within the structure of the Gene Ontology. Since the Gene Ontology is divided into three categories, we can generate separate graphs for Molecular Functions (MF), Cellular Component (CC), and Biological Process (BP), respectively. Since Combined Graphs can still be large and difficult to navigate, more accessible representation of the functional data is obtained by bar charts. Bar charts create a transversal cut of the GO graph at the selected Ontology level. In this example, we choose to generate a bar chart at level 7 to get a broad overview of the functional distribution in this dataset.
Summary of Statistics Charts
The functional annotation module contains many additional visualization tools to graphically summarize annotation results.
Some of the most useful ones are:
Measured Times and Cloud Unit Consumption
- Blast via CloudBlast against Viridiplantae: 2.5 hours / 130k Cloud Units
- InterProScan via CloudInterProScan: 3.5 hours / 100k Cloud Units
- EggNOG-Mapper: 20 min
- GO Mapping and GO Annotation: 10 min
Results
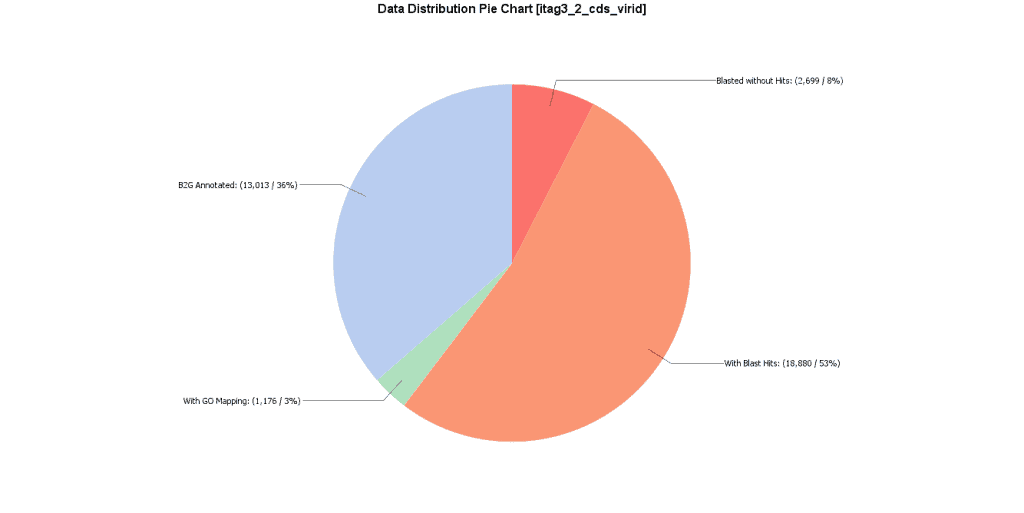
From a total of 35,768 CDS sequences, 79.5% could be annotated with high-quality GO terms as a combination of the 3 employed annotation strategies. InterProScan contributed protein domain or family information for 87% and GO annotations for 56% of the sequences. EggNOG-Mapper annotated 89% with orthologous groups or other information, and 45% with GO annotations. 77.5% of the sequences could be related to GO terms through the Blast2GO methodology. A part from that, enzyme codes could be assigned to 23.5% of the sequences.
CloudBlast, CloudInterProScan and eggNOG annotation can be run at the same time, which reduces the overall analysis time down to roughly 4 hours.
References
- Conesa A., Götz S., Garcia-Gomez JM., Terol J., Talon M. and Robles M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics (Oxford, England), 21(18), 3674-6.
- Conesa A. and Götz S. (2008). Blast2GO: A comprehensive suite for functional analysis in plant genomics. International journal of plant genomics, 2008, 619832.
- Götz S., Garcia-Gomez JM., Terol J., Williams TD., Nagaraj SH., Nueda MJ., Robles M., Talon M., Dopazo J. and Conesa A. (2008). High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic acids research, 36(10), 3420-35.