Time-Course Differential Expression Analysis
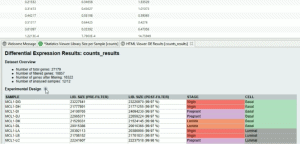
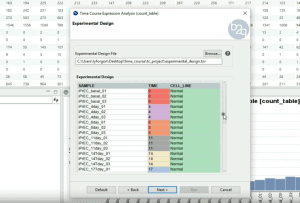
The Time Course Expression Analysis tool allows performing a differential expression analysis of expression data arising from a time course RNA-seq experiment. This application is based on the maSigPro Bioconductor package, which implements a two-step regression strategy to detect genes with significant temporal expression changes and significant differences between experimental groups. This video shows the analysis of count data coming