Pairwise Differential Expression Analysis
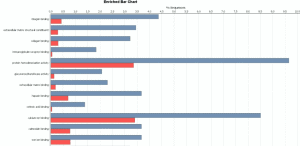
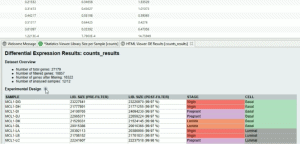
The Pairwise Differential Expression Analysis tool is designed to perform differential expression analysis of count data arising from an RNA-seq experiment. The application, which is based on the software package “edgeR”, allows the identification of differentially expressed genes between two experimental conditions by applying quantitative statistical methods. This video shows the performance of a pairwise differential expression analysis in which