Specific tools for a new generation of sequencing
Third Generation Sequencing (TGS) is getting a notable increase in relevance by surpassing some of the limitations of NGS technologies. In that sense, the main feature of TGS is the capacity to produce much longer reads than NGS. These long reads facilitate the assembly of complex genomes and the study of alternative RNA splicing.
However, TGS long reads still present a high sequencing error rate that can negatively affect downstream analysis. Quality assessment and control tools (such as LongQC) have become essential in every long reads data analysis to overcome this issue.
Quality Control of Long Reads data
Several tools have been proposed to perform this step in the last few years. However, all of them must face the significant differences between data produced by the different technologies (mainly, PacBio and Nanopore) and the nature of the sequenced molecule (DNA or RNA).
LongQC is a bioinformatic tool that allows the quality control of long-read data from major TGS technologies. It provides useful statistics and charts to evaluate the quality of any dataset efficiently. Moreover, LongQC does not need reference genomes to perform its analyses, so it is particularly valuable before conducting a de novo genome assembly. Lastly, LongQC is compatible with both PacBio and Nanopore data and allows both genomic and transcriptomic data analysis.
LongQC use to assess the quality of Long Reads datasets
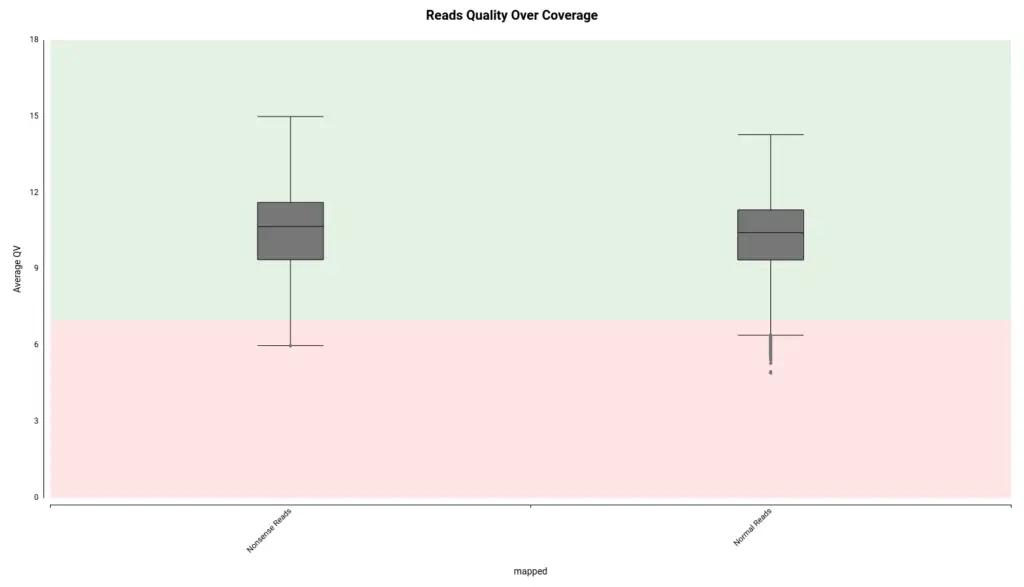
Among them, the authors consider the coverage module as the pipeline’s core. This module generates coverage statistics and plots by calculating the overlap between the reads using a modified version of minimap2. The coverage module detects non-sense reads that could be contaminants or artifacts from the sequencing process. In that logic, non-sense reads proportion can accurately estimate the quality of the sequencing data.
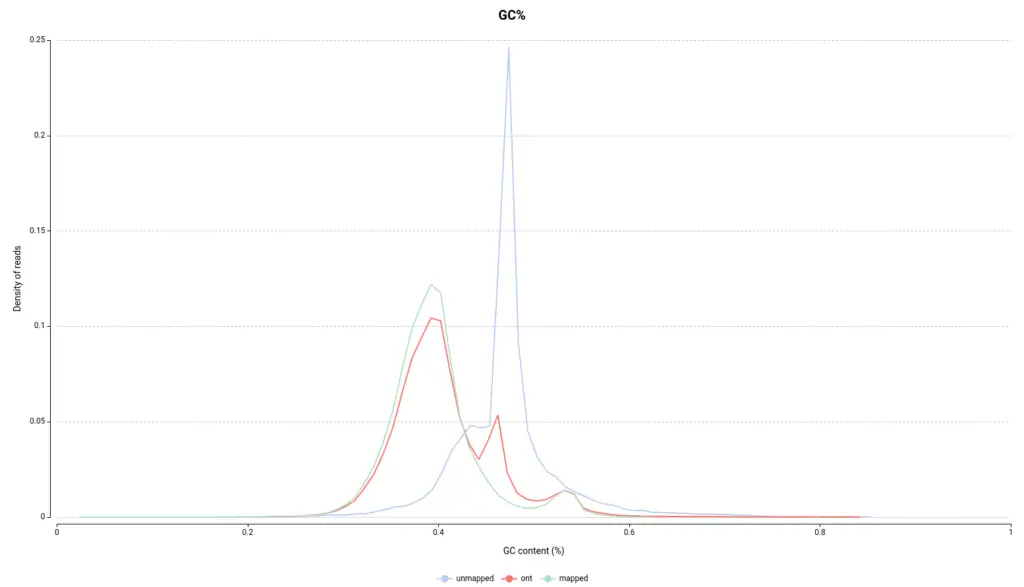
The pipeline also includes more “classic” methods, such as the analysis of GC content, the per-base quality values, or the distribution of the read sequences length. Most of them are very similar to some modules included in FastQC, the widely standardized quality control tool for NGS.
LongQC in OmicsBox
- LongQC is included in the General Tools Module along with some other quality control and preprocessing utilities such as FastQC or Trimmomatic.
- Launching a LongQC analysis in OmicsBox is straightforward and allows quickly adjusting the analysis execution to the specific data characteristics.
- The OmicsBox implementation allows the simultaneous analysis of more than one sample. This feature can compare the results of analyzing several files from the same experiment.
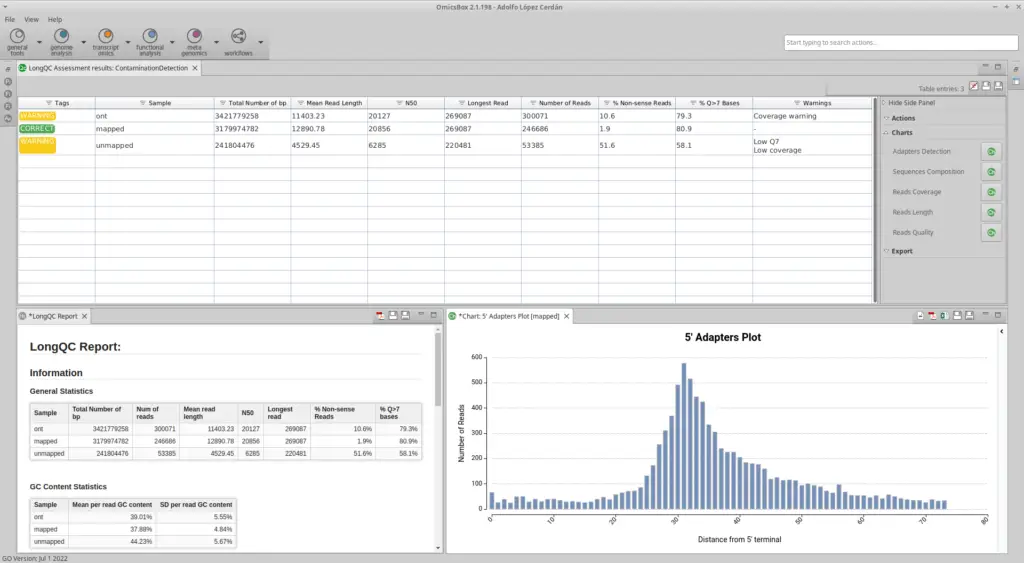
- As a result of the LongQC run, OmicsBox shows a table with some useful general statistics and an Html report with a more extensive set of quality metrics.
- The user can display a wide range of descriptive plots from the results table. Moreover, the user can pick the samples to display in the charts.
- Having all this information, the user can decide to apply additional preprocessing steps to the data or directly go on different stages of long reads data analysis, such as genome assembly by Flye or transcriptome preprocessing by SQANTI3.
Check out our mini-tutorial:
References
LongQC: A Quality Control Tool for Third Generation Sequencing Long Read Data.
Yoshinori Fukasawa, Luca Ermini, Hai Wang, Karen Carty, Ming-Sin Cheung.
G3: Genes, Genomes, Genetics, 10(4): 1193-1196, 2020
Useful Links