Unlocking Long Read Insights with OmicsBox
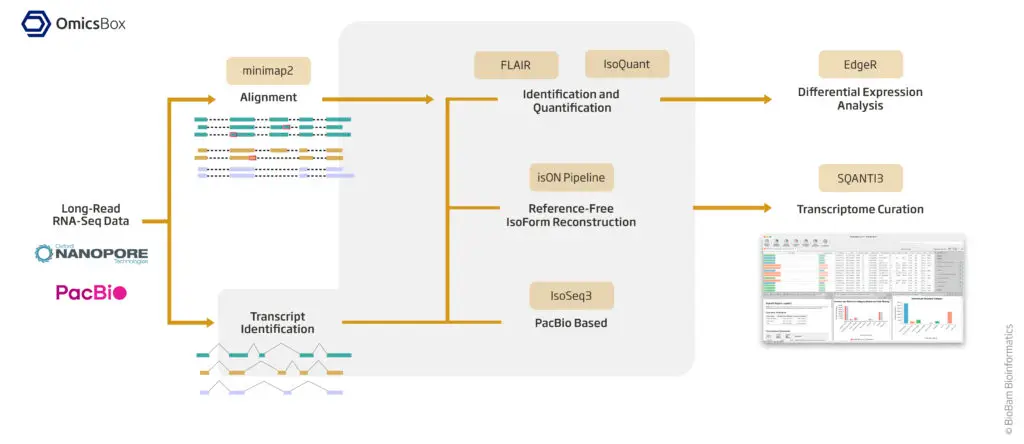
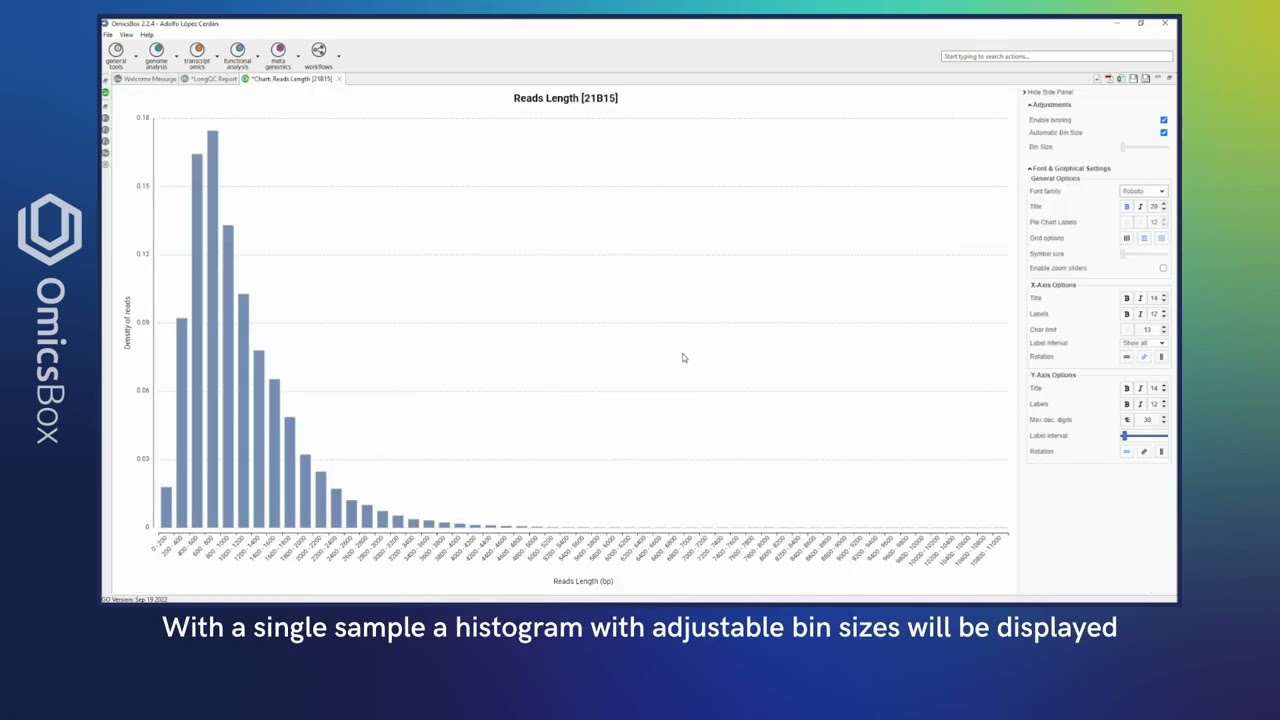
Long Read technologies (mainly PacBio and Oxford Nanopore) have been proven to be optimal for characterizing and analyzing isoforms, as each read can contain an entire transcript. With OmicsBox set of long reads tools you will be able to accurately reconstruct a transcriptome, assess the quality, and quantify isoforms easily.
Features
- minimap2
Use minimap2 to align your long reads. Minimap2 has many different parameters, but in OmicsBox, you can use its predefined preset of options to adapt the tool to your dataset and align it accurately.
- IsoSeq3
With IsoSeq3, you will be able to obtain corrected and polished long reads from PacBio subreads or ccs. IsoSeq3 is a pipeline composed of different tools. However, with OmicsBox, you just have to upload your subreads or ccs PacBio bam files and adjust the desired parameters.
- FLAIR
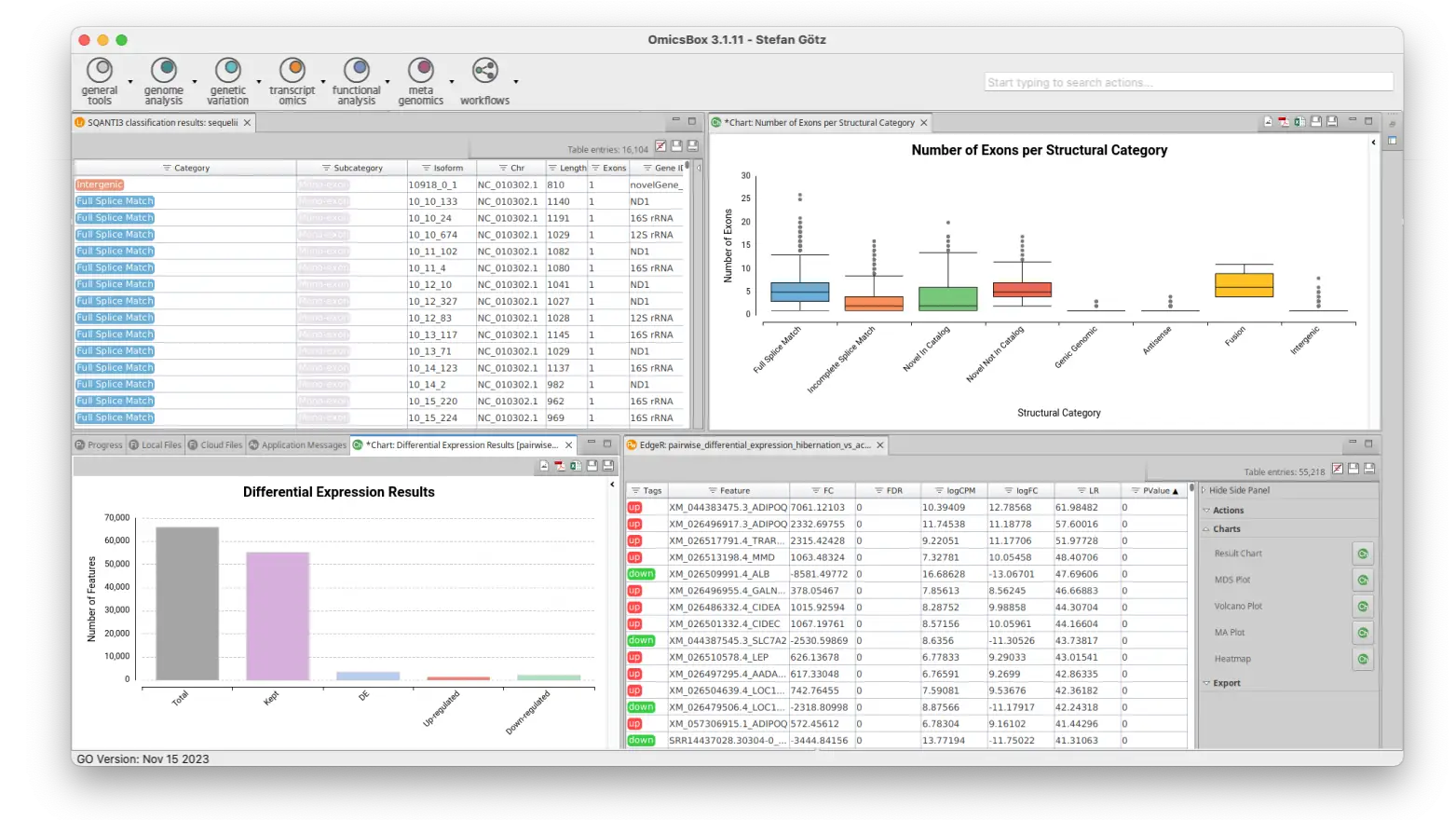
FLAIR is a tool that constructs a transcriptome from long reads. It is reference-guided, which means that you need a reference genome and splice junction information (short reads or a reference annotation). Using this tool, you will receive a well-defined transcriptome in addition to a transcript count table. This enables downstream differential expression analyses for experiments with multiple samples.
- IsoQuant
IsoQuant is a tool for the genome-based analysis of long RNA reads, such as PacBio or Oxford Nanopore. Similarly to FLAIR, IsoQuant also reconstructs a transcriptome from long reads. Nevertheless, IsoQuant can be used just with the reference genome and not splice junction information and it can perform quantification of reference transcripts and genes, without transcript model construction.
- isON PIPELINE
Use this pipeline to perform a reference-free transcriptome reconstruction using a long reads dataset. If you are working with non-model species whose transcriptomes and genomes are not well-defined, this pipeline is optimal for you. This pipeline consists of three steps: clustering, correction, and reconstruction, each with different options that can be easily executed in OmicsBox.
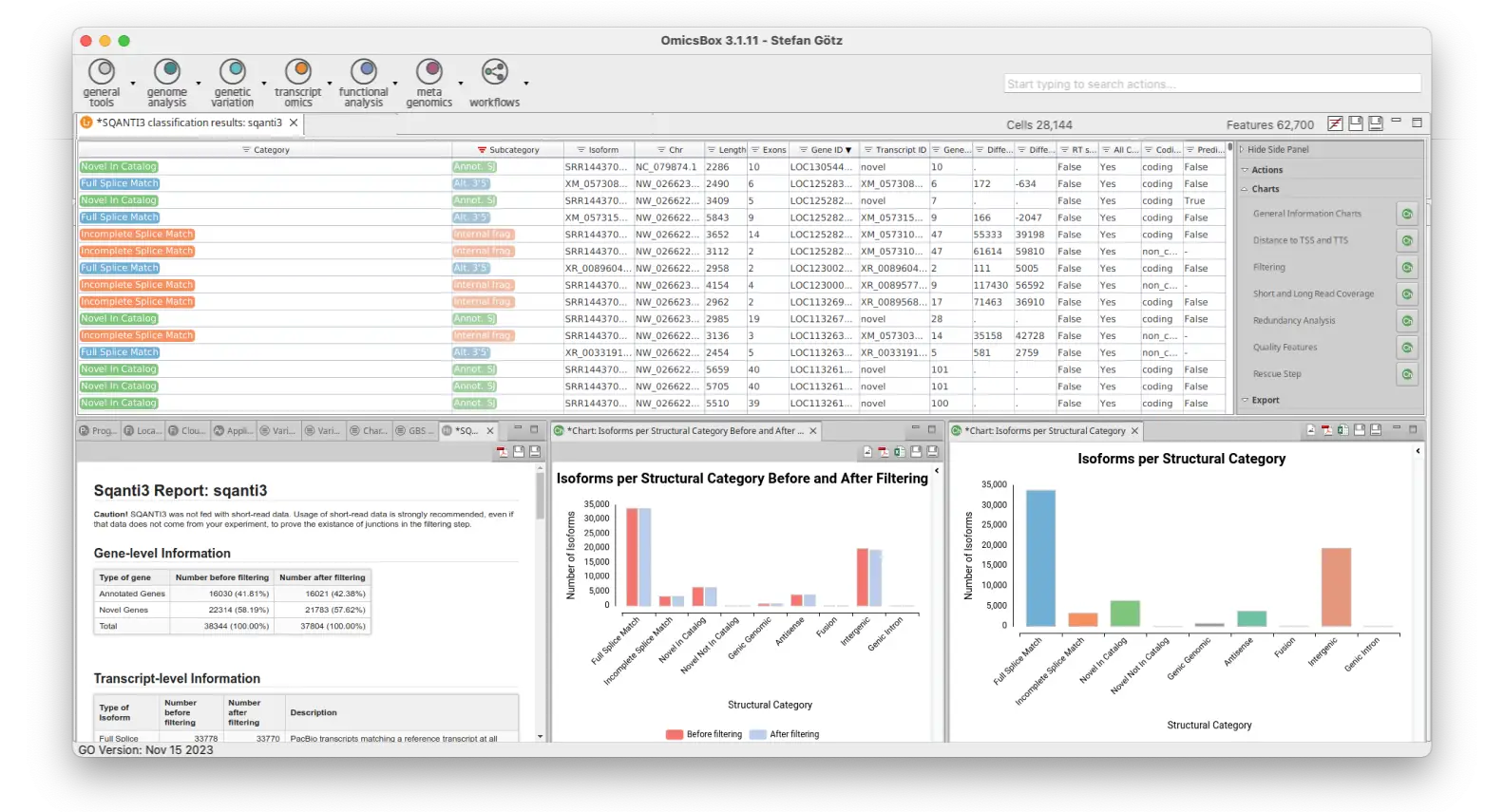
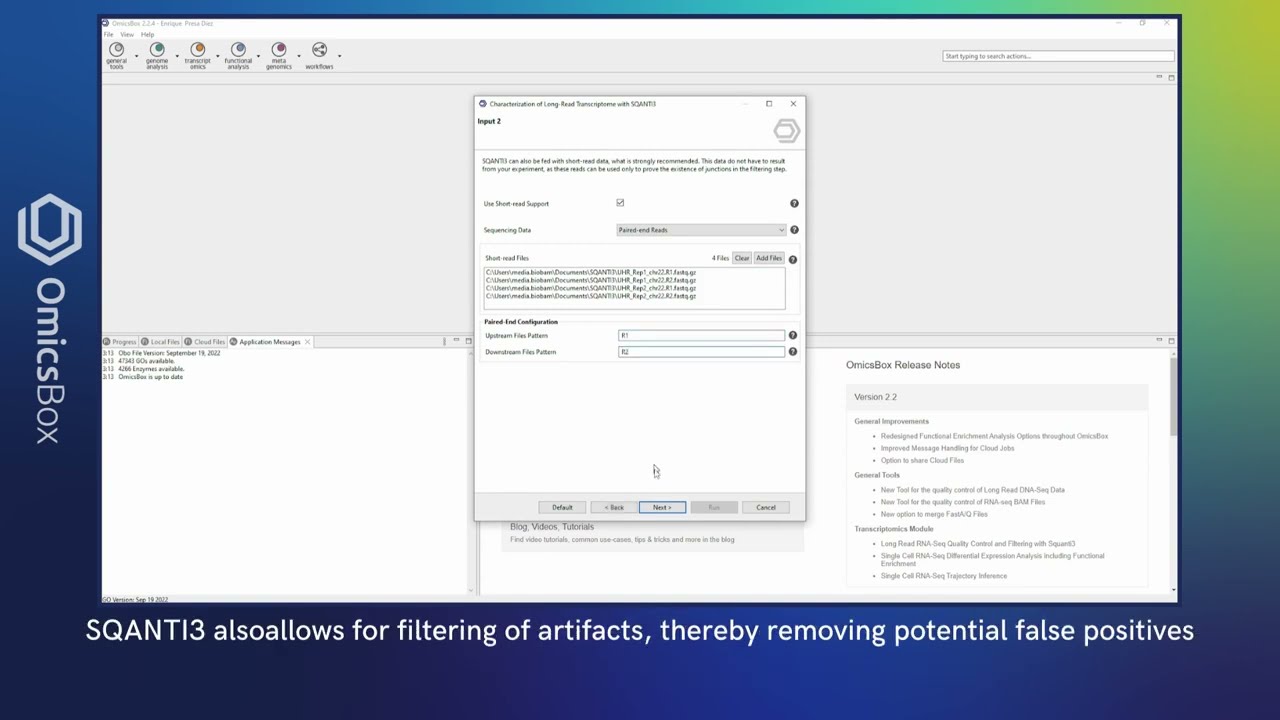
- SQANTI3
Once you have your long-reads transcriptome, you can use SQANTI3 to assess its quality and characterize each isoform within your transcriptome model. Additionally, SQANTI3 can be used to filter out false positive isoforms to obtain a curated model.
With the Long-Reads set of tools of OmicsBox, you can perform end-to-end long-read analyses, no matter whether you have a lot of information about your organism of study or not. One of the most vital features in this module is the powerful transcriptome characterization and quality control through SQANTI3, which provides a wide variety of rich and insightful visualizations. In addition, you will be able to conduct differential expression analysis with long reads after generating a transcriptome using FLAIR or IsoQuant.
Get Started with a Free Trial
Get familiar with all new Modules and Features
OmicsBox works out of the box on any standard PC or laptop with Windows, Linux and Mac.
Videos, Tutorials & More
Keep reading about Long Reads

Long Read Sequence Alignment with Minimap2
Sequence alignment is an essential element of genomics research, playing a significant role in aiding scientists in deciphering the intricacies of genetic…
Decoding long Read Sequenced Transcriptomes: Flair vs. Stringtie 2
Transcriptome reconstruction is a challenging bioinformatic problem. The development of long-read sequencing technologies has made it easier to solve this issue…