A BioBam scholarship supported project – with OmicsBox.
Background and Project Overview:
Garlic (Allium sativum L.) is one of the most important remunerative bulbous spices and medicinal crops grown commercially. It is rich in protein, phosphorus, potash, calcium, magnesium and carbohydrates. The freshly peeled garlic cloves contain 62.8% moisture, 29% carbohydrate, 6.3% protein, 1% mineral matter, 0.8% fiber, 0.1% fat, 1% total ash, 0.03% calcium, 0.31% phosphorus, 0.0001% iron, nicotinic acid and vitamins whose metabolic pathways have not been worked out at genomic level. In the present study, paired-end reads were generated using Illumina Hiseq 2000 sequencing technology.
Assembling of the sequencing data resulted in contigs being obtained. Differentially expressed genes were identified and characterized between the tissues of leaf and clove. Besides, greater emphasis was laid on the genes, which were highly expressed in clove, later it is assumed to contain a high content of bioactive compounds. Further analysis resulted in the identification of genes that are conceivably involved in the organosulfur compounds, phenolic compounds, nitrogen, carbohydrate, lipid and other bioactive metabolic pathways.
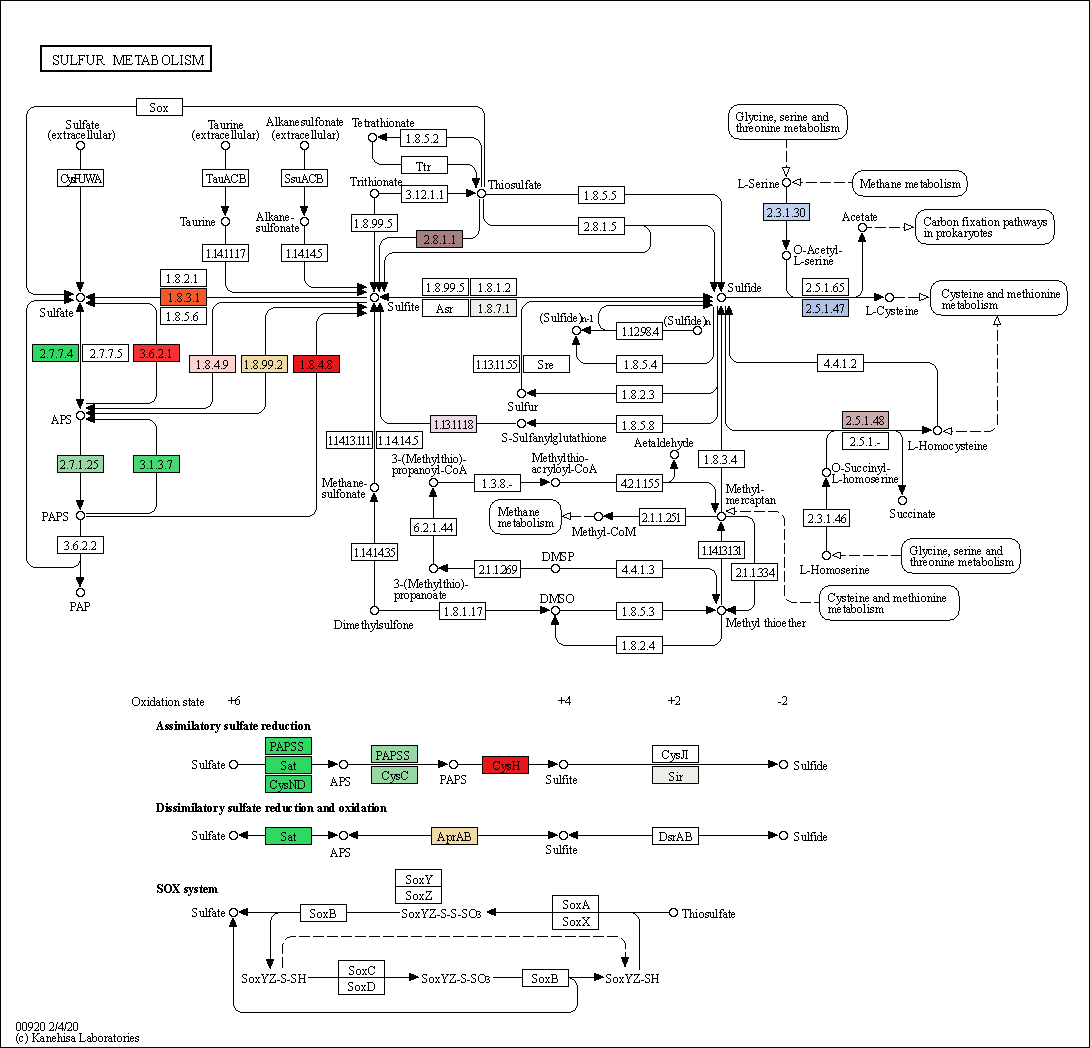
Figure. Sulfur metabolism pathway in garlic transcriptome generated by KEGG.
Contribution of OmicsBox
OmicsBox was involved in most of the bioinformatics analysis. Transcriptome and functional analysis were commonly used in this study. Initially, quality assessment of raw sequences was performed using FASTQ quality inspection tools and sequences were processed using the OmicsBox pre-processing tools. The Functional Analysis module was used for the functional gene annotation process. Almost all the functional annotation tools like BLAST, InterProScan annotation, Gene Ontology analysis (mapping and annotations), Enrichment analysis, Metabolic pathway analysis (KEGG annotations) and EggNOG annotations were useful for this study. Functional enrichment analysis helped to identify the specific genes that were present in the garlic. The cloud facility provided by OmicsBox was convenient to annotate these large transcriptome sequences.
This project has been supported with free access to all OmicsBox resources, including all modules.
Researchers:
- Malyaj R Prajapati: ORCID: https://orcid.org/0000-0002-8542-9732
- Jitender Singh: ORCID: https://orcid.org/0000-0003-0715-1365
- Pankaj Kumar
Affiliation:
College of Biotechnology, SVPUA&T, INDIA