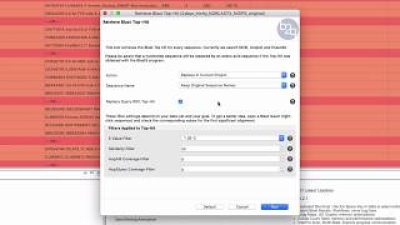
RNA-seq data is sometimes difficult to match with proteins, due to the short length of the reads. When this is the case, it might be useful to try to find EST hits, which can then be used to find new protein matches. In this demo, we will show how to retrieve top EST hits and the different options that this tool offers.
In this video, we use an RNA-seq dataset in which many sequences did not find a Blast result. To further characterize sequences without result, we first show how to select and extract sequences to a new project. Then, we perform a local Blast against an EST database and finally we explain in detail how to retrieve the top EST hits. Using the top EST hits as queries, we perform a Blastx search in order to find new protein matches and improve functional characterization with OmicsBox/Blast2GO.