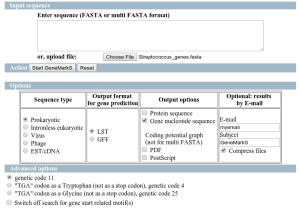
How to change sequence names coming from Prokaryotic GeneFinding
When running GeneFinding the sequences receive a name with the predicted genes. The first part of the sequence identifier comes from the genome reference sequence name (de-novo assembly) and then a _orfx is appended, where x is a number. Sometimes this name is not useful to proceed with downstream analysis or compare results from other experiments. Is there any way