At PAG33, we had the opportunity to participate once again in one of the most important international meetings for Plant and Animal Genomics. In addition to our exhibition booth, this year we were pleased to organize an industry workshop focused on practical non-code bioinformatics workflows using OmicsBox. The session brought together both methodological overviews and real research applications, and we were honored to host two guest speakers, Amit Dhingra and Javier Terol. They are both leading researchers in their fields, using OmicsBox as part of their routine data analysis.
The workshop aimed to demonstrate how integrated, user-friendly bioinformatics software can support researchers in handling complex omics datasets, from raw sequencing data to biologically meaningful results.
OmicsBox Overview and Integrated Workflows
The workshop opened with an introduction by our CEO, Stefan Götz, who presented OmicsBox, our comprehensive desktop application designed to cover a broad range of bioinformatics analyses within a single environment. OmicsBox includes five different modules: functional annotation, transcriptomics, genomics, and genetic variation, which cover complete analysis pipelines from raw sequencing reads down to biological insights.
This is why OmicsBox is used by research laboratories worldwide, as well as by biotech companies, to analyze diverse types of omics data without requiring command-line expertise.
Functional Annotation and Transcriptomics in Non-Model Organisms
The workshop continued with Stefan explaining the functional and transcriptomics pipelines, which are designed to work with virtually any species, including non-model and novel organisms.
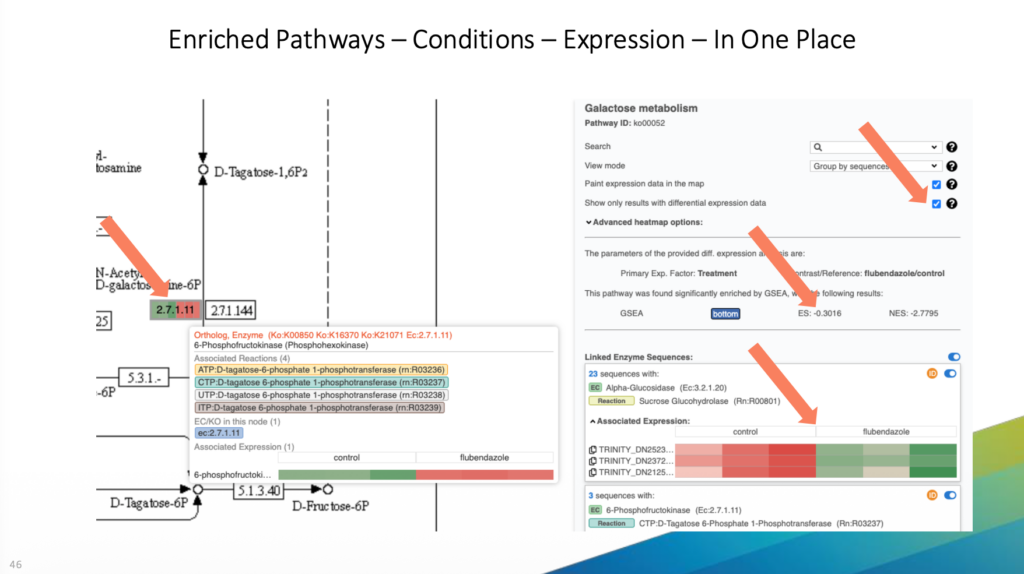
Using a real use case, Stefan demonstrated a complete workflow starting with raw FASTQ reads from a novel organism, Ascaridia galli, for which no reference transcriptome was available at that time. The analysis covered the generation of a de novo transcriptome, its functional annotation and pathway identification, and differential expression analysis. A detailed explanation of the analysis pipeline is available in this webinar.
The example illustrated how researchers can move from just sequencing reads to the identification of differentially expressed pathways between experimental conditions.
Large-Scale Genetic Variation Analysis with Cloud Parallelization
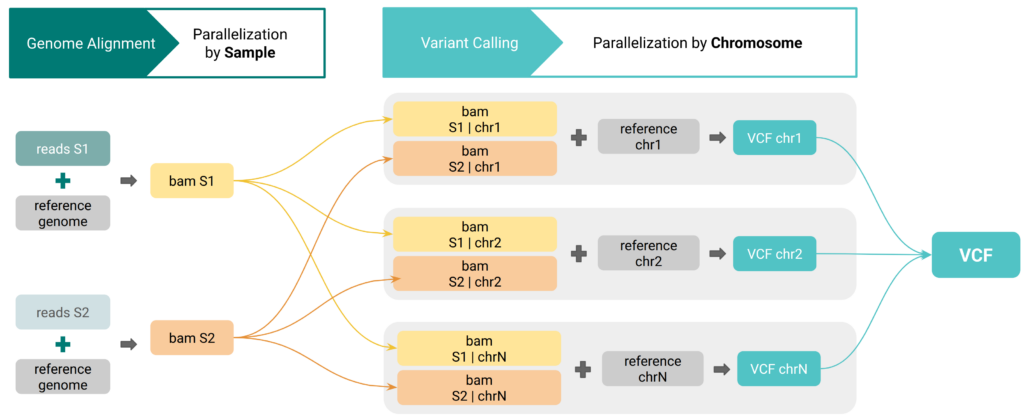
The second technical block, presented by Marta, focused on the Genetic Variation module and the challenges associated with analyzing datasets that include a large number of samples.
Handling high-volume sequencing data often requires substantial computational resources and careful pipeline configuration. Marta showed how OmicsBox addresses these challenges through automatic cloud-based parallelization, which operates in the background. Users do not need to configure infrastructure or manage resource allocation manually.
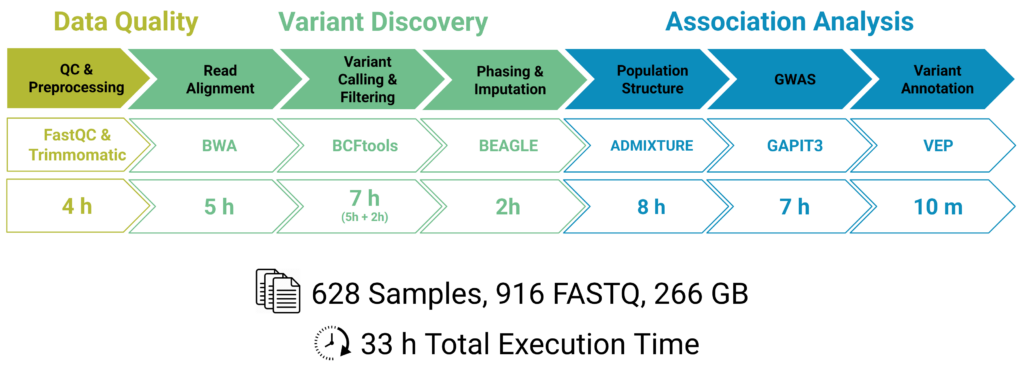
This approach was demonstrated using a large Vitis vinifera dataset, consisting of more than 600 samples. Despite the dataset size and complexity, the complete variant analysis pipeline was completed in 33 hours, illustrating how scalable computing can significantly reduce turnaround times while keeping the workflow accessible to non-specialist users.
The complete analysis of the dataset and the obtained results are explained in more detail in this webinar.
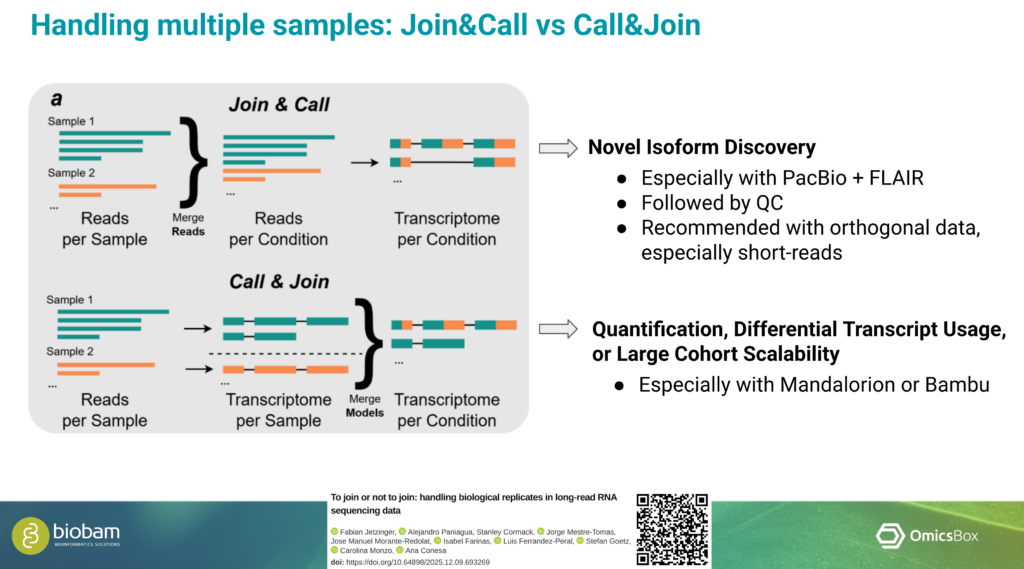
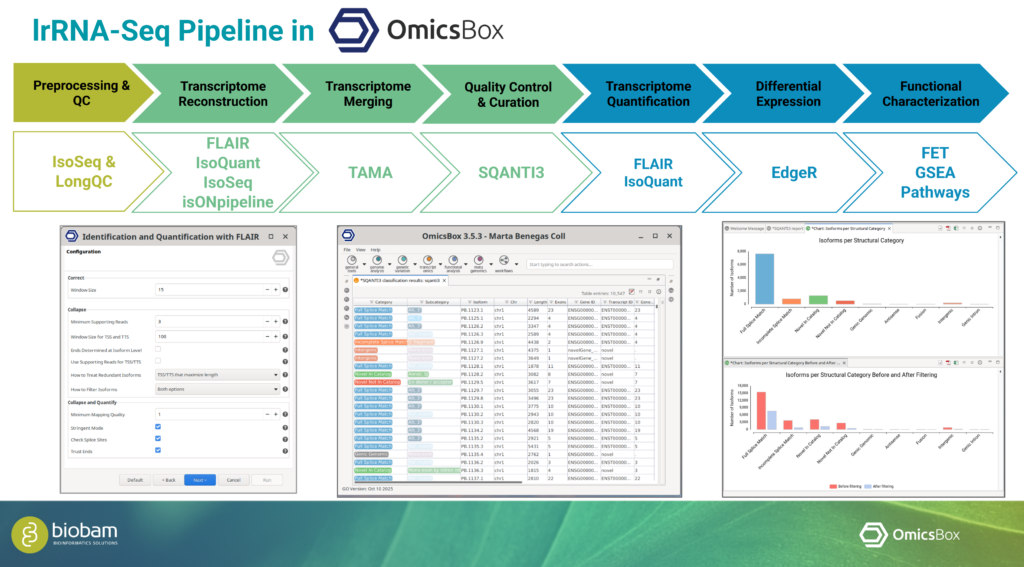
Considerations for Long-Read RNA-Seq Data Analysis
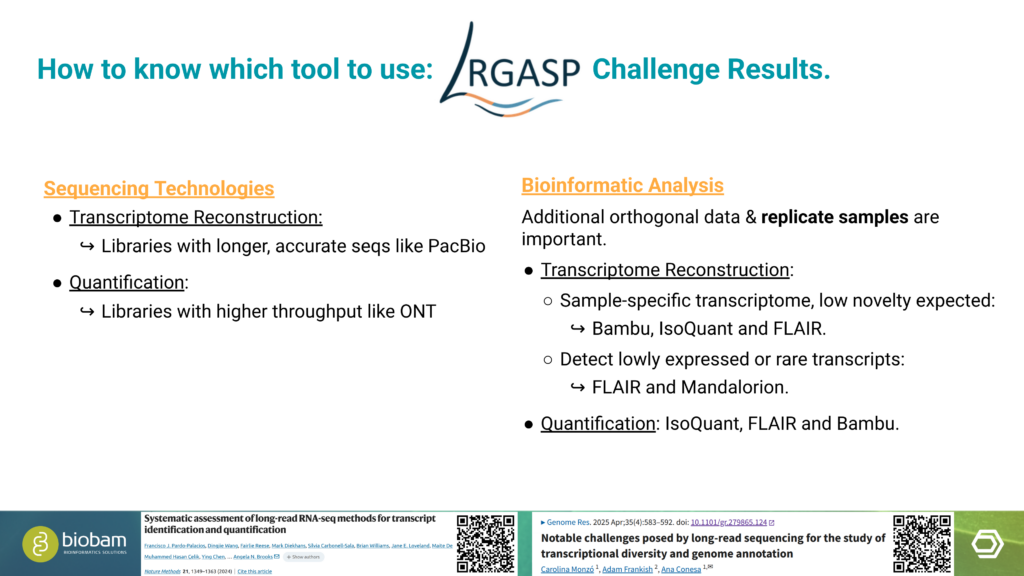
In an additional talk, Marta shared practical insights into the analysis of long-read RNA-Seq data. She emphasized that the choice of sequencing technology, analysis tools, and pipeline design should be driven by the characteristics of the data and the biological objectives of the study. Careful planning at these stages is essential to obtain reliable and informative results.
OmicsBox supports long-read data generated by both Oxford Nanopore Technologies (ONT) and PacBio platforms. The software also provides multiple tools for transcriptome characterization, enabling flexible workflows that can be adapted to different experimental designs and research questions. This adaptability makes it well-suited for diverse long-read transcriptomics use cases. This was demonstrated by analyzing two very different datasets, generated from hexaploid sweet potato and a bear.
Real Research Applications
A key component of the workshop was the contribution from our guest researchers, Amit Dhingra and Javier Terol, who presented how they apply OmicsBox to their own horticulturalresearch projects. Their presentations highlighted how integrated analyses across different data types can support biological interpretation and have contributed to several of their published studies. These real-world examples illustrated how OmicsBox fits into ongoing research and supports the generation of publishable results.
Some example publications from Amit Dhingra and Javier Terol’s research labs using OmicsBox include:
Comparative transcriptomic analyses of citrus cold-resistant vs. sensitive rootstocks might suggest a relevant role of ABA signaling in triggering cold scion adaption. Primo-Capella, A., et al. BMC Plant Biol 22, 209 (2022). Comparative transcriptomic analyses of citrus cold-resistant vs. sensitive rootstocks might suggest a relevant role of ABA signaling in triggering cold scion adaption
Transcriptome analysis reveals activation of detoxification and defense mechanisms in smoke-exposed Merlot grape (Vitis vinifera) berries. Hewitt, S., et al. Scientific Reports 14, 21330 (2024). Transcriptome analysis reveals activation of detoxification and defense mechanisms in smoke-exposed Merlot grape (Vitis vinifera) berries
Impact of heat stress, water stress, and their combined effects on the metabolism and transcriptome of grape berries. Hewitt, S., et al. Sci Rep 13, 9907 (2023). Impact of heat stress, water stress, and their combined effects on the metabolism and transcriptome of grape berries
Metatranscriptomic analysis of tomato rhizospheres reveals insight into plant-microbiome molecular response to biochar-amended organic soil. Hewitt, S., et al. Frontiers in Analytical Science, (2023). Frontiers | Metatranscriptomic analysis of tomato rhizospheres reveals insight into plant-microbiome molecular response to biochar-amended organic soil
Concluding Remarks
The PAG33 industry workshop provided an opportunity to showcase not only the technical capabilities of OmicsBox but also its practical value in day-to-day research. Key aspects emphasized throughout the session included:
- Cloud connectivity that enables scalable analyses.
- A graphical, user-friendly interface.
- Interactive result exploration, facilitating data interpretation.
Together with the contributions from Amit and Javier, the workshop demonstrated how integrated bioinformatics tools can help researchers move efficiently from raw data to biological insight in real research scenarios.
We thank all participants for their interest and engagement, and we look forward to continuing the discussion at future events.
About the Author
 Marta Benegas
Marta Benegas studied biotechnology at the Valencia Polytechnic University (UPV) and continued her studies with a Master's in Bioinformatics at the Autonomous University of Barcelona (UAB), Spain. After her master's degree, she started her professional career at Biobam where she is now working as a bioinformatics specialist and support manager.
At the moment she is mainly focused on Single-Cell technologies developing various pipelines which allow getting from reads to functional insights at a single-cell resolution. These developments are available in OmicsBox, BioBam’s software solution.
Marta Benegas
Marta Benegas studied biotechnology at the Valencia Polytechnic University (UPV) and continued her studies with a Master's in Bioinformatics at the Autonomous University of Barcelona (UAB), Spain. After her master's degree, she started her professional career at Biobam where she is now working as a bioinformatics specialist and support manager.
At the moment she is mainly focused on Single-Cell technologies developing various pipelines which allow getting from reads to functional insights at a single-cell resolution. These developments are available in OmicsBox, BioBam’s software solution.