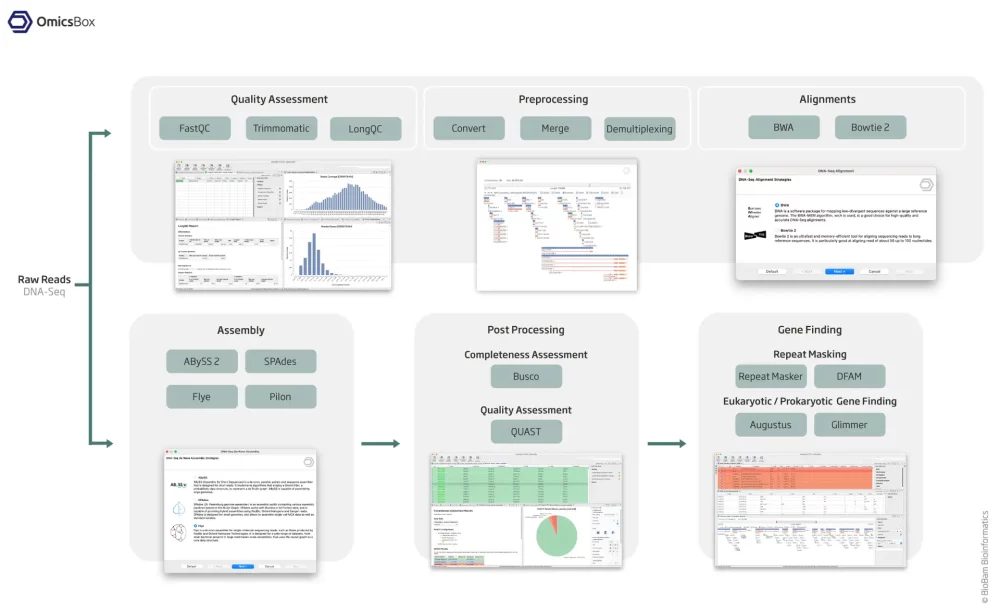
Quality Control And Assessment
Use FastQC and Trimmomatic to perform the quality control of your samples, to filter reads and to remove low quality bases.
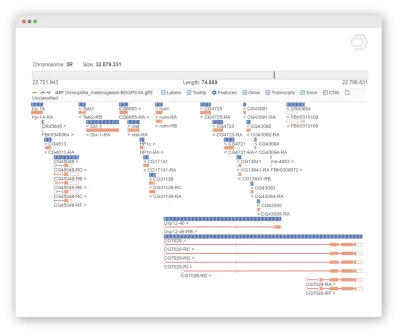
Genome Browser
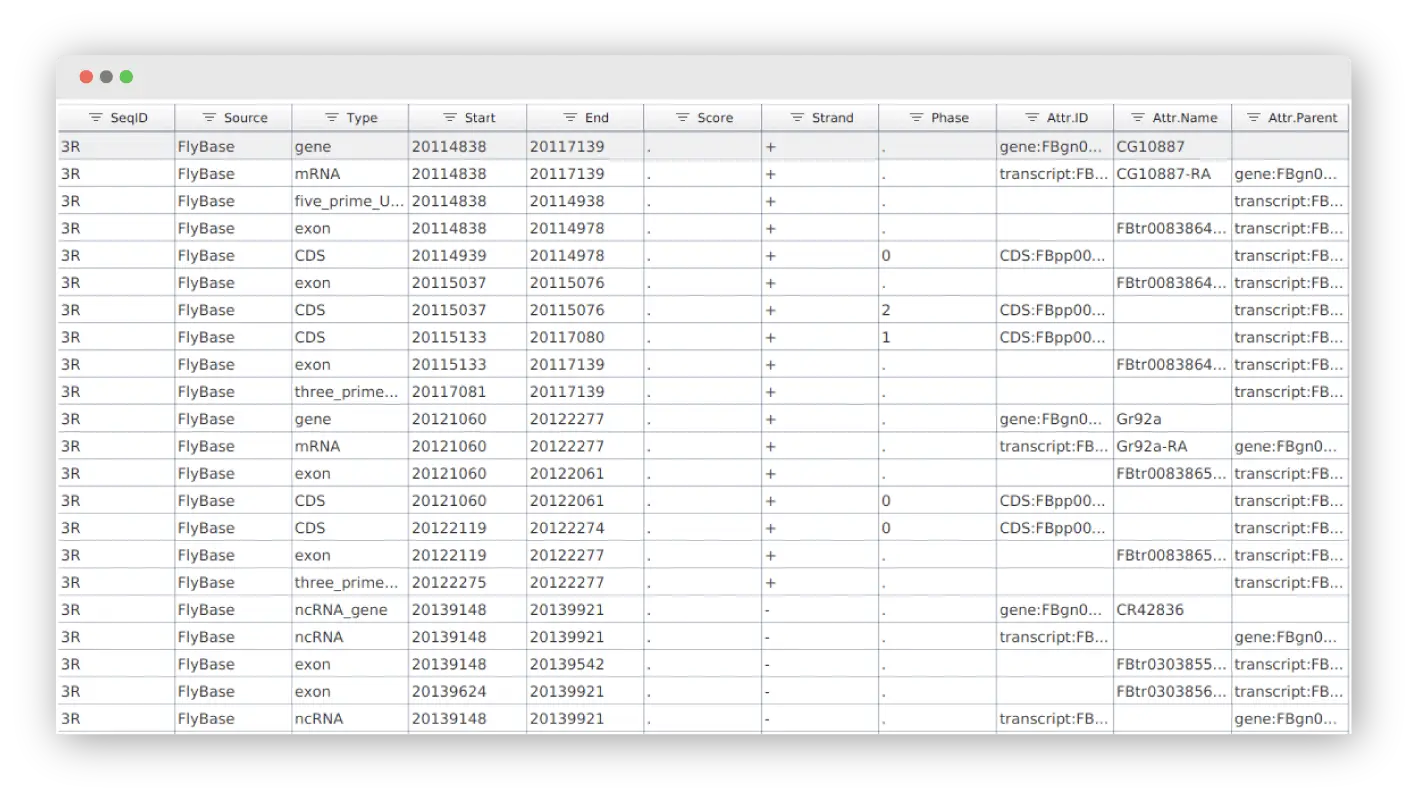
Visualize your annotations in form of tracks to combine the genome sequences (.fasta) with alignments (.bam), intron-exon structure (.gff) and variant data (.vcf).
De-Novo Assembly
The assembly feature allows to reconstruct whole genome sequences without a reference genome or specific hardware requirements. Assemble sequencing data from both, short and long read technologies with 3 different algorithms: ABySS, SPAdes and Flye.
Repeat Masking and Gene Finding
Mask repeats of genome assemblies with RepeatMasker to improve downstream gene predictions.
Perform prokaryotic (Glimmer) and eukaryotic (Augustus) gene predictions to characterize genome structure.
Alignment and Polishing
Align short sequencing reads against large sequences with BWA or Bowtie 2, and correct draft assemblies from long reads with Pilon.
Multi-Locus Sequence Typing (MLST)
Characterize bacterial isolates unambiguously. This procedure considers the alleles present in (usually) seven well-characterized housekeeping genes.
- DNA-Seq de novo assembly with ABySS 2
- DNA-Seq de novo assembly with SPAdes
- DNA-Seq de novo assembly with Flye
- DNA-Seq alignment with BWA
- DNA-Seq polishing with Pilon
- Repeat masking with RepeatMasker
- Evidence-based eukaryotic gene predictions with Augustus
- Prokaryotic gene finding with Glimmer
- Genome Assembly Completeness Assessment with Busco
Dive in Genome Analysis: Our Module Highlights
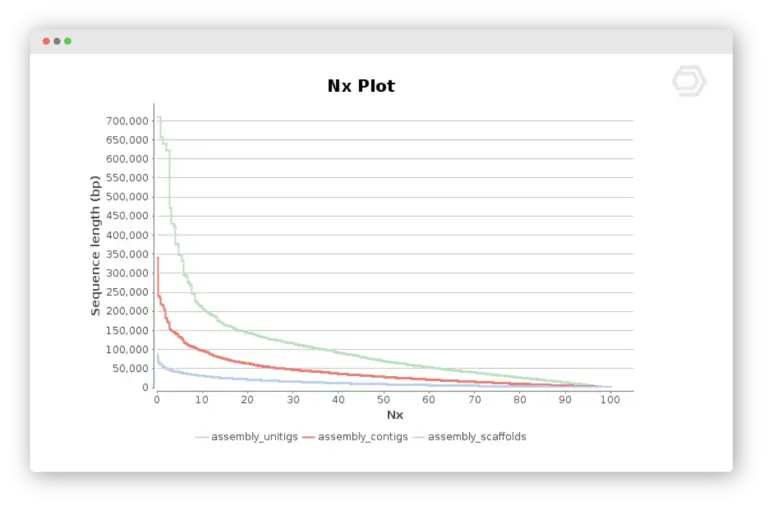
Quality Assessment and comparison of de novo genome assemblies using QUAST
Understanding the quality and reliability of genome assemblies is a critical step in any genomics workflow.QUAST, a widely recognized tool for assembly evaluation, is seamlessly integrated into the Genome Analysis module of OmicsBox. By comparing multiple de novo assembly strategies, users can examine key quality metrics, including contiguity, completeness, accuracy, and fragmentation.
This guided walkthrough demonstrates how OmicsBox enables researchers to make informed decisions about the optimal assembly strategy for their datasets, streamlining genomic analysis with precision and clarity.
Get Started with OmicsBox
Get familiar with all new Modules and Features with a Free Trial or a Custom Demo.
OmicsBox works out of the box on any standard PC or laptop with Windows, Linux and Mac.
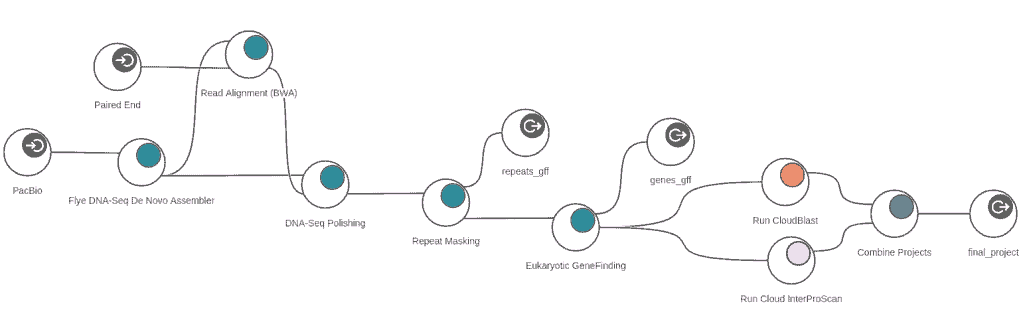
Workflows

Eukaryotic Genome Analysis
Generate a genome draft of eukaryotic species by assembling DNA-Seq reads without additional prior information. Detect and mask repetitive sequences and improve the gene prediction by providing RNA-Seq data.
Prokaryotic Genome Analysis
Genomes of bacteria and other prokaryotic organisms can be assembled and characterized in a fast and sensitive way. Proceed with the functional annotation of the resulting gene sequences.