Our team member and PhD student Fabian Jetzinger recently contributed to the organization of the LongTREC summer school, held at the Earlham Institute in Norwich, UK. The 4-day course, organized by the MSCA doctoral network LongTREC welcomed 27 participants from across Europe for an intensive introduction to bioinformatic analyses of long-read transcriptomics data.
A diverse range of complex topics was covered during the course: mapping, transcriptome reconstruction, quality control, differential expression, functional analysis, single-cell sequencing, epitranscriptomics, and metatranscriptomics.
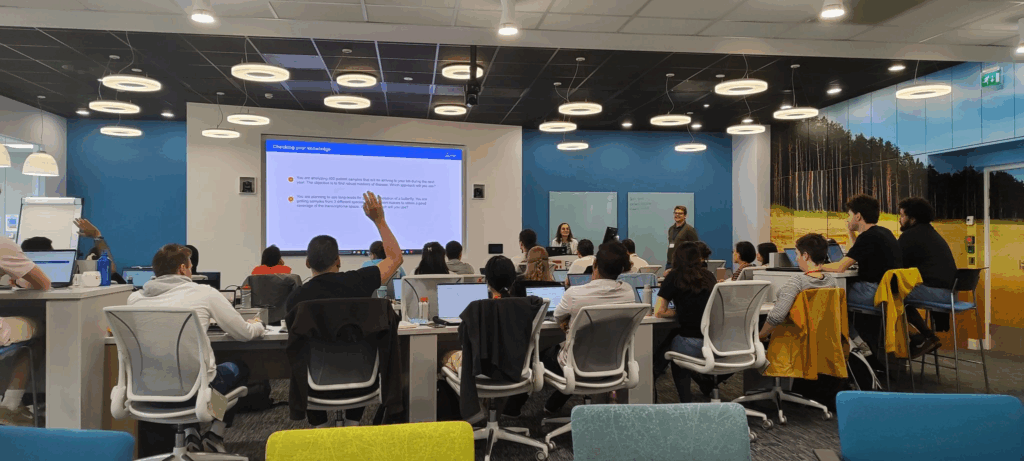
Fabian held presentations on transcriptome reconstruction and quantification, transcriptome quality control and curation, and functional analysis. The attendees, which were made up of master’s students, pre- and post-doctoral researchers, and even senior bioinformaticians, were highly engaged, leading to lively discussions and knowledge-sharing throughout the week.
“I am the only person in my lab working with long-read sequencing data. It’s challenging to keep up with these fast-evolving technologies. The course was incredibly useful to gain an overview of the field and get advice from experts,” commented one attendee.
Beyond the lectures, LongTREC students were invited on a guided tour of the Earlham Institute’s state-of-the-art laboratory facilities. Although they specialize in bioinformatics, the opportunity to see sequencing technology such as the PacBio Revio system firsthand was an inspiring and memorable experience.
Materials created for the LongTREC summer school are publicly available in a github repository. Fabian can be reached at fjetzinger@biobam.com for questions and further discussion.

About the Author


